This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers find mechanism that ensures correct DNA segregation in cell division
A research team led by Dr. Gary Ying Wai Chan from the School of Biological Sciences at The University of Hong Kong (HKU), has uncovered a new mechanism that ensures correct DNA segregation in cell division, where improper cell division will lead to the development of cancer.
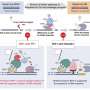
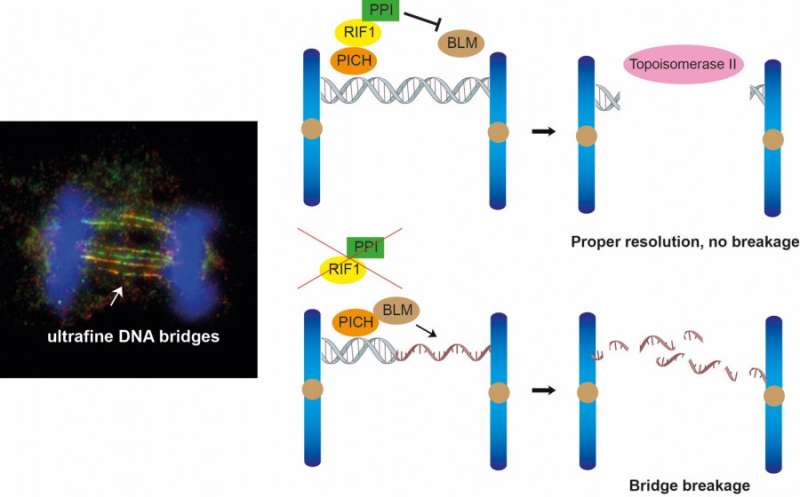
The team's findings, published in the journal Cell Reports, focus on the roles of two proteins, RIF1 and protein phosphatase 1 (PP1), in resolving ultrafine DNA bridges. These bridges are formed when sister chromatids are connected by DNA joint molecules during mitosis. If these DNA bridges cannot be resolved or removed properly, they will eventually break and cause DNA damage in the daughter cells, which can lead to the development of cancer cells.
A human life begins with a single cell—the fertilized egg. This single cell needs to replicate and divide into approximately 37 trillion cells. The process by which a cell replicates its DNA then equally segregates into two identical cells is known as mitosis, which is a vital process for growth and replacing worn out cells. However, equal segregation of DNA is one of the most challenging tasks in mitosis.
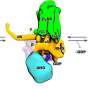
Our DNA is organized into 23 pairs of chromosomes, each of which is replicated into two sister chromatids. During mitosis, these sister chromatids are separated into two identical daughter cells. They are often connected by DNA joint molecules, which can form long fine DNA threads known as ultrafine DNA bridges when the two sister chromatids are separating under a pulling force. If these DNA bridges cannot be resolved or removed properly, they will eventually break, causing damage in the daughter cells. In a worst-case scenario, this DNA damage can potentially lead to the development of cancer.
To elucidate the detailed mechanism of how cells resolve the DNA bridges, the team led by Dr. Gary Ying Wai Chan uncovered the roles of two proteins, RIF1 and protein phosphatase 1 (PP1) in regulating the resolution of ultrafine DNA bridges.
The role of RIF1 and PP1 in resolving DNA bridges
In 2007, two research groups led by Professor Erich Nigg and Professor Ian Hickson discovered the existence of ultrafine DNA bridges by identifying the first two ultrafine bridge-binding proteins, PICH and BLM. Later, RIF1 was also identified as a bridge-binding protein. It is known that PICH is the first protein recognizing the DNA bridges, then it recruits BLM and RIF1 to ultrafine DNA bridges; however, the exact mechanism by which these bridge-binding proteins work to resolve the bridges remains unclear.
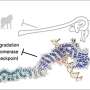
To understand the role of RIF1 in resolving DNA bridges, the research team employed a revolutionary genome editing technology, the CRISPR/Cas9, to deplete RIF1 in a cell model during mitosis. During which, the team found that loss of RIF1 leads to increased formation of DNA damage and micronuclei due to breakage of ultrafine DNA bridges, as our data suggest that RIF1 plays a crucial role in preventing double-stranded DNA bridges from converting into single-stranded DNA, which is more susceptible to breakage.
RIF1 was found to achieve this by recruiting PP1, a protein phosphatase, which reduces the interaction between BLM and PICH and thereby reduces the amount of BLM on the bridges, which the team discovered that is responsible for unwinding the bridge DNA to single-stranded DNA.
Finally, the double-stranded DNA bridges protected by RIF1-PP1 are believed to be resolved by another enzyme known as topoisomerase IIα, which could mediate double-stranded DNA decatenation to ensure proper DNA segregation.
This research not only sheds light on the novel regulatory mechanism by which RIF1-PP1 facilitates the resolution of DNA bridges, but also reveals how ultrafine DNA bridges can induce DNA damage and genome instability if they are not properly resolved.
The findings suggest that DNA bridge-binding proteins may serve as potential therapeutic targets for the development of anticancer drugs, as DNA bridges are considered a source of genome instability that drives tumorigenesis.
"The discovery that RIF1 recruits PP1 to the bridges is the first step in understanding how the resolution of ultrafine DNA bridges is regulated by protein phosphorylation/dephosphorylation," said Dr. Gary Chan. "The next step is to identify the target substrates of the RIF1-PP1 complex, which can advance our understanding of how different bridge-binding proteins interact with each other and may lead to the identification of new therapeutic targets for cancer."
More information: Nannan Kong et al, RIF1 suppresses the formation of single-stranded ultrafine anaphase bridges via protein phosphatase 1, Cell Reports (2023). DOI: 10.1016/j.celrep.2023.112032
Journal information: Cell Reports
Provided by The University of Hong Kong