This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Detecting rapidly mutating bacteria and viruses with AutoPLP
As we now know from our experience with the COVID-19 pandemic, the microbes responsible for some infections can rapidly mutate into variants that evade detection and treatment.
But now, researchers reporting in ACS Infectious Diseases have developed a procedure that could help researchers catch up to these sneaky pathogens. Their "AutoPLP" technique designs nucleic acid probes to detect new variants quickly, accurately and easily.
Many diagnostics, such as those based on the polymerase chain reaction (PCR) detect pathogens by analyzing genetic material. Rolling circle amplification (RCA) works in a similar way, though an advantage is that it doesn't involve the complex temperature cycling that PCR does.
Both techniques require nucleic acid probes with sequences matching those of the target pathogen in specific locations, but RCA uses highly specific "padlock probes" (PLPs). As a pathogen mutates, its genetic sequence changes as well, and researchers have to keep redesigning their probes.
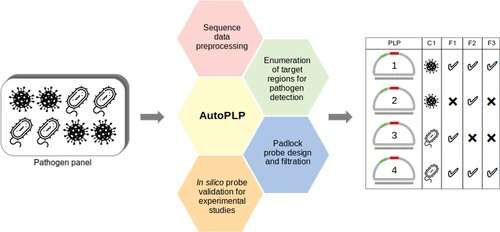
So, Sowmya Krishnan, Ruben Soares, M. Michael Gromiha and Narayanan Madaboosi wanted to create a tool that could not only design these PLPs automatically, but also, for the first time, systematically consider all the necessary technical parameters at once to make the entire process easier and more robust.
Their tool, a computer program called "AutoPLP," was named after the PLPs it designs. The program can take the genome sequences of similar pathogens as input and run a series of analyses and database searches, outputting a set of customized PLP sequences. The team used the program to design probes against the rabies virus, a virus transmitted between animals and people, and Mycobacterium tuberculosis, the bacterium responsible for tuberculosis, and compared them to previously reported ones.
For the rabies virus, AutoPLP targeted three genes, yielding probes with a higher and narrower range of melting temperatures than those in the literature. For M. tuberculosis, they designed a total of 13 probes specifically targeting two genes responsible for drug-resistant strains with the program. The researchers say that this tool could help hasten the discovery of new pathogen variants, helping combat them rapidly and effectively via precise molecular diagnostics.
More information: Sowmya Ramaswamy Krishnan et al, AutoPLP: A Padlock Probe Design Pipeline for Zoonotic Pathogens, ACS Infectious Diseases (2023). DOI: 10.1021/acsinfecdis.2c00436
Journal information: ACS Infectious Diseases
Provided by American Chemical Society