Leveraging the structure of bacterial host cell receptors to detect cancer
Bacteria in our bodies bind to various host cell surface receptors, which determines where the bacteria live and how they behave. These receptors, made up of chains of sugar molecules called glycans, are more than meets the eye. Cells existing within progressive disease states like cancer can have an increased number of glycan receptors on their surfaces.
The lab of Tina Iverson, Louise B. McGavock Chair and professor of pharmacology, uncovered the structural mechanism by which streptococcus bacteria bind to host cell glycans, opening the door to new ways of using bacterial molecules to potentially detect cancer cells.
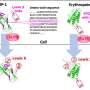
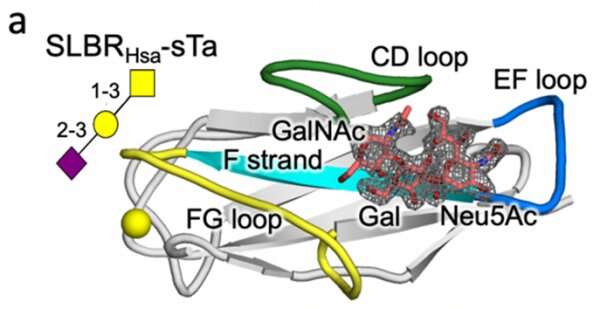
Previous research has shown that bacteria such as streptococci use adhesive molecules called SLBRs to bind to glycan receptors on the surface of host cells. The Iverson lab hypothesized that these bacteria may adapt the structure of their SLBRs to bind to a wide range of host glycan receptors to help them inhabit new niches in the body.
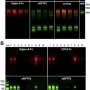
Turning to protein engineering, the Iverson lab determined which SLBR structural elements control the choice of host cell receptor binding. By artificially modifying the DNA coding for SLBRs and purifying the resulting proteins from bacteria, they discovered that three loops in the SLBR—called CD, EF and FG—control which glycans the bacteria will bind to.
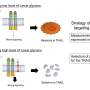
Bacterial attachment to host cell receptors is the first step in infection. Understanding the mechanism behind this process may also help researchers identify cells enriched in surface glycans that exist within disease states.
As part of this study, the Iverson lab found that commensal bacteria of the mouth bind to a sugar-coated or glycosylated protein of the mouth that is also overrepresented in highly aggressive cancer cells in most carcinomas. By developing techniques that alter SLBR glycan selectivity, Iverson is determined to tackle a new long-term goal: developing reagents that can identify highly aggressive cancers. The glycans that cells express may serve as biomarkers to help physicians target diseases for therapeutic intervention.
"We are working to expand a library of glycan binding agents we developed in this study to allow us to detect a greater range of glycans," said Iverson, also a professor of biochemistry. "Eventually we would like to develop it into diagnostic kits."
The study "Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation" was published in Nature Communications in May 2022.
More information: Barbara A. Bensing et al, Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation, Nature Communications (2022). DOI: 10.1038/s41467-022-30509-y
Journal information: Nature Communications
Provided by Vanderbilt University