A simple method to determine phase preference of proteins on live cell membranes
National University of Singapore scientists have demonstrated a simple and fast method to determine if a biomolecule partitions into lipid domains on live cell membranes.
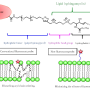
Cellular membranes are dynamic assemblies of lipids and proteins with some components organized as domains. Proper cell function requires the partitioning of lipids and proteins into these domains, which are often rich in cholesterol and sphingolipids. However, they are too small (10 to 200 nm in size) and dynamic (possibly only tens of milliseconds in lifetime) to be observed even with modern super-resolution microscopy techniques. Traditional methods to determine domain localization involve biochemical assays which require many cells, are prone to artifacts as they are conducted in vitro, and are slow. Although fluorescence-based techniques can probe these domains in live cells, they require specialized instrumentation and are often difficult to interpret.
The research team led by Professor Thorsten Wohland from the Departments of Biological Sciences and Chemistry, National University of Singapore (NUS) developed a simple fluidizer-based method to determine if a molecule prefers to partition into lipid domains on cell membranes.
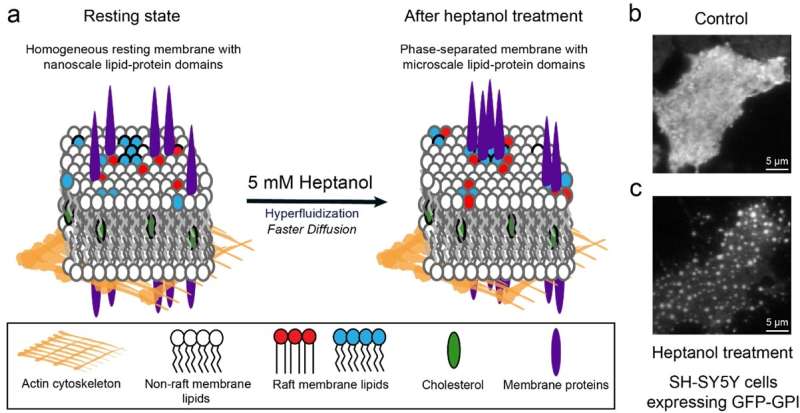
The team added heptanol to live cells and showed that within 15 minutes it induces clustering of the nanometer-sized lipid domains into larger micrometer-sized domains that are easily detectable by standard fluorescence microscopes. The method works with both molecules that are genetically labeled with fluorescent proteins and those labeled using extrinsic labels, for example, antibodies.
Prof. Wohland said, "The phase preference of molecules used to be difficult and time-consuming to establish. This new method, detected by chance, provides results in at most 15 minutes on live cells and can essentially be seen by eye in a simple microscope."
The team hopes that this technique will enable a quick and facile identification of domain localization and will aid the wider research community.
More information: Anjali Gupta et al, Heptanol-mediated phase separation determines phase preference of molecules in live cell membranes, Journal of Lipid Research (2022). DOI: 10.1016/j.jlr.2022.100220
Journal information: Journal of Lipid Research
Provided by National University of Singapore