Study reveals structure of receptor implicated in type 2 diabetes and more
Researchers from the University of Southern California, Merck & Co., Skoltech, MIPT, UCLA, and the Université de Sherbrooke have determined the structure of the human leukotriene B4 receptor 1, involved in inflammatory, infectious, allergic, and tumorigenic diseases. Published in Nature Communications, the analysis of the structure reveals how the receptor recognizes its binding partners and interacts with them. This opens up avenues for designing better drugs that would target the receptor to treat Type 2 diabetes and other pathologies.
Receptors are the protein-based equipment cells use to receive and transmit signals. A receptor becomes activated when it binds a messenger molecule called an agonist, whereupon it relays the signal, which regulates some biological function. Antagonists, by contrast, shut down the receptor when bound. Agonists and antagonists are collectively known as ligands.
The human leukotriene B4 receptor 1, or hBLT1, regulates inflammation-related processes—such as the recruitment of T cells—as well as the proliferation and migration of smooth muscle cells. That receptor has been associated with diseases, including asthma, influenza, arthritis, atherosclerosis, diabetes, and cancer.
Since its discovery in 1997, there have been a number of attempts to develop hBLT1 ligands for use as drugs, but they had many side effects, low efficacy, and the body took comparatively long to eliminate them. A likely explanation for this is that the hBLT1 ligands used are not specific to that receptor and engage in other unwanted interactions. Learning more about the structure of the receptor and how it binds ligands can allow pharmacologists to design better, more selective drugs.
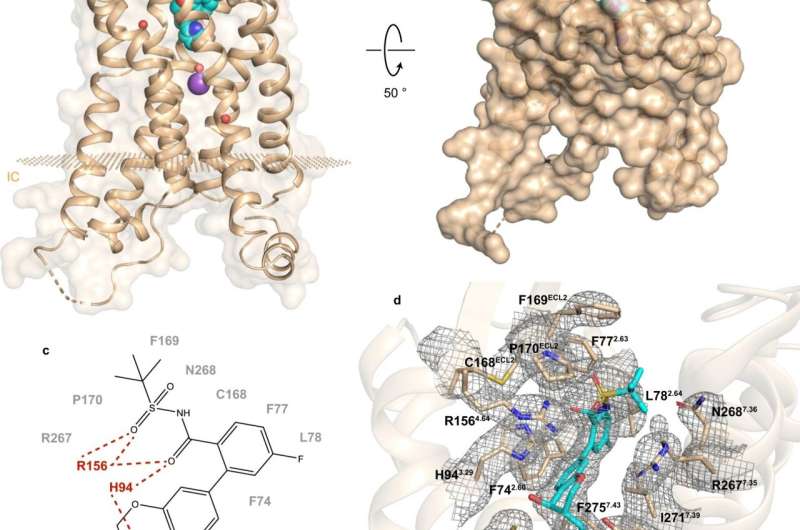
A recent study by a Russian-U.S.-Canadian collaboration sheds light on the makeup and functioning of hBLT1. Vadim Cherezov, professor of Chemistry at USC and the head of the MIPT Laboratory for Structural Biology of GPCRs, commented: "We have determined the 2.9-angstrom-resolution crystal structure of the hBLT1 receptor in complex with a selective antagonist, MK-D-046, developed by Merck & Co. This structure should help to rationally design better therapeutics to treat type 2 diabetes and other inflammatory conditions."
Structure determination was complemented by site-directed mutagenesis and docking studies—an experimental and a computational method, respectively. According to Skoltech Assistant Professor Petr Popov, "this made it possible to reveal the key determinants of intermolecular interactions between the receptor and the ligands."
The analysis of hBLT1 structure reveals how the receptor recognizes and binds ligands, suggesting a putative ligand access channel buried in the receptor's membrane. More specifically, the findings hint at the possible ways the receptor might bind its endogenous agonists. That is, compounds naturally produced by the body to bind to that receptor and activate it.
By improving our understanding of hBLT1 structure and functioning, the study opens up possibilities for structure-based drug design.
More information: Nairie Michaelian et al, Structural insights on ligand recognition at the human leukotriene B4 receptor 1, Nature Communications (2021). DOI: 10.1038/s41467-021-23149-1
Journal information: Nature Communications
Provided by Skolkovo Institute of Science and Technology