Screening for macrocyclic peptides
Macrocyclic peptides are promising candidates for pharmaceuticals, but their screening is difficult. Scientists have now developed an easy-to-use, high-throughput screening assay for cyclic peptides with affinity to ubiquitin, a protein that helps to degrade proteins and induce cell death. The results could lead to novel drug candidates against cancer, according to the study published in the journal Angewandte Chemie.
Drugs based on peptides (small proteins) are often too large to pass through cell membranes. To make such peptides more compact and stable—and thus more efficient—researchers are investigating their closed versions, called macrocyclic peptides. Pharmaceuticals based on macrocyclic peptides are interesting candidates for the modulation of regulatory proteins, which is an important goal in cancer research.
However, screening for such peptides is difficult. "Well-established screening approaches often require highly expensive instrumentation and appropriate training," says Ashram Brik from the Technion-Israel Institute of Technology, Israel, who is set up to develop alternative strategies.
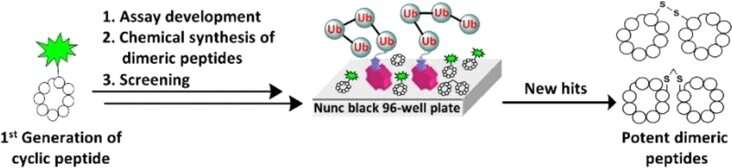
The researchers designed a simple screening assay based on competitive binding. Competitive binding means that unknown peptides are tested for their capabilities to displace a known peptide with known binding affinity to a target. The readout is simply the change in fluorescence, which is provided by the fluorescence label attached to the known peptide. The researchers say that that this setup enables fast, high-throughput, cheap, and easy-to-implement screening of peptide libraries.
The researchers used the assay to screen for peptides binding strongly to ubiquitin, a small cellular protein that cells use to tag dysfunctional proteins and mark them for degradation. They identified several peptide candidates with high binding affinity, which could be further tweaked to make them more functional.
"For example, we subjected the peptides to chemistries such as alkylation arylation, oxidation and dimerization, the first step to make polycyclic peptides," Brik said. "Polycyclic peptides have added value crucial to be considered as drug candidates." Scientists assume that cyclization makes the peptides more stable than their open analogs, and their compact shapes help them to enter cells more easily.
The authors prepared dimers of cyclic ubiquitin-binding peptides, where two identical cyclic peptides are linked, and screened them again for their affinity to ubiquitin. They discovered strongly binding candidates and tested them for their regulatory role in live cancer cell lines. The authors found the dimeric cyclic peptides entered the cells easily and induced cell death, and they were more potent than the control peptide.
In addition, the authors suggest that the novel circular peptides could be labeled and used as staining agents for ubiquitin with antibody-like properties.
More information: Ganga B. Vamisetti et al, The Development of a Fluorescence‐Based Competitive Assay Enabled the Discovery of Dimeric Cyclic Peptide Modulators of Ubiquitin Chains, Angewandte Chemie International Edition (2020). DOI: 10.1002/anie.202013392
Journal information: Angewandte Chemie International Edition , Angewandte Chemie
Provided by Wiley