Researchers identify post-translational formation of strained cyclophanes in bacteria
NUS researchers have identified post-translational modifying enzymes in bacteria that can create a new family of cyclic peptides for new therapeutics.
Cyclic peptides are a well-known class of molecules that possess a range of biological activities including anti-bacterial, immunosuppressant and anti-cancer properties. The challenge to creating new types of cyclic peptides lies in the discovery of new catalytic reactions by chemical synthesis or by using enzymes. One strategy to accomplish this involves carbon-carbon (C-C) bond formation. However, C-C bond-forming reactions at unactivated carbon-hydrogen (C-H) bonds are difficult because these positions are inert, and the selectivity is not easy to control. Overcoming these barriers will allow the creation of a new range of cyclic peptides in an efficient and predictable manner.
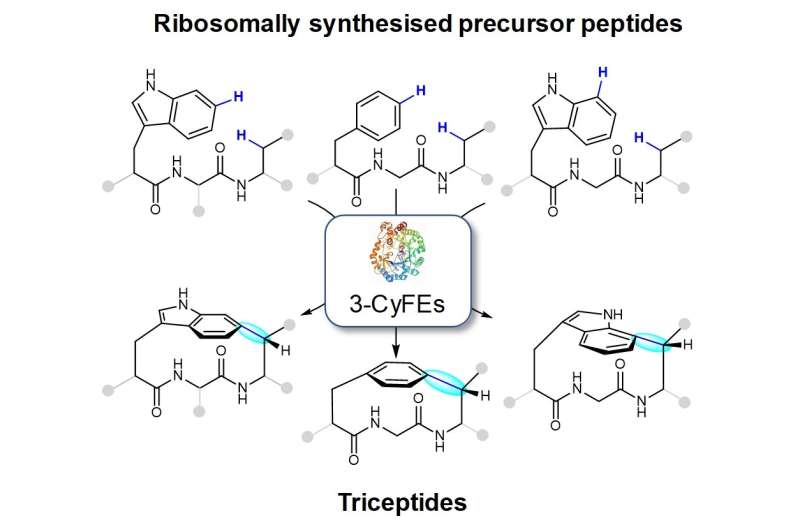
The research team led by Prof Brandon Morinaka from the Department of Pharmacy, NUS has found reactions catalyzed by a suite of post-translational modifying enzymes from various bacteria which can potentially be used to develop important drug molecules. These enzymes can catalyze the formation of a C(aryl)-C(sp3) bond between the side chains of two amino acids. This molecular arrangement is the defining feature for a class of natural products named triceptides (three residues in cyclophane peptides) (see figure). The ring-closing reaction results in the formation of strained peptide cyclophanes. Although these enzymes are encoded in several hundred bacterial genomes, their function and their end products have remained largely unknown.
These enzymes offer a new strategy for the modification of peptides and can be applied to generate therapeutic peptides that display unique binding to molecular targets. The cyclisation of peptides is a general strategy used in nature and synthetic chemistry to prevent degradation by digestive enzymes. The prevention of degradation is an essential attribute necessary for drugs to reach their target and represents a hurdle to using peptides as drugs.
These enzymes are able to install the rings in a variety of patterns with different amino acids. This means that the breadth of products is potentially large and can be applied to diverse targets in drug discovery. Diverse bacteria from a range of microbiomes were found to encode the cyclophane forming enzymes. The diversity of products hints that peptide cyclophanes are more prevalent than previously known and may confer a general use or function among this class of natural products.
The team plans to produce additional triceptide natural products and to understand the enzyme mechanism. Their long-term goals are to generate a diverse range of peptide cyclophane products which can be used as a starting point for targeting diseases of interest.
Prof Morinaka said, "Post-translational cyclophane forming enzymes offer a new method for creating diverse cyclic peptides that provide new opportunities in peptide engineering and drug discovery."
More information: Nguyen TQN et al, Post-translational formation of strained cyclophanes in bacteria, Nature Chemistry DOI: 0.1038/s41557-020-0519-z Published 2020.
Journal information: Nature Chemistry
Provided by National University of Singapore