Decoding messages in the body's microscopic metropolises
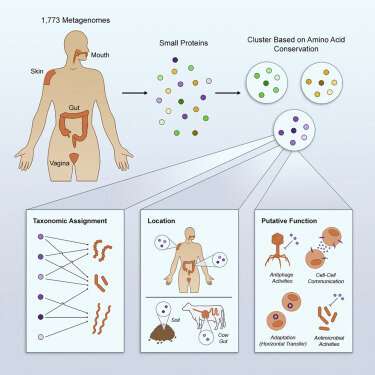
A study aimed at identifying and examining the small messenger proteins used by microbes living on and inside humans has revealed an astounding diversity of more than 4,000 families of molecules – many of which have never been described previously.
The research, led by Stanford University and now published in Cell, lays the groundwork for future investigations into how the trillions of bacteria, archaea, and fungi that compose human microbiomes compete for resources, attack and co-exist with one another, and interact with our own cells.
"Because it is much more difficult to search for sequences encoding small proteins than it is to trawl for large proteins, our comprehension of the small proteins expressed by microbial communities has always been lacking," said Nikos Kyrpides, a Berkeley Lab senior scientist who contributed to the work.
Yet, very small proteins made of 50 or fewer amino acids, which can move though cell walls and membranes, perform many essential tasks that mediate an organism's interactions with the environment.
These functions, combined with the fact that they are easier to synthesize and manipulate than large molecules, make small microbial proteins a potential source of new medicines.
More information: Hila Sberro et al. Large-Scale Analyses of Human Microbiomes Reveal Thousands of Small, Novel Genes, Cell (2019). DOI: 10.1016/j.cell.2019.07.016
Provided by Lawrence Berkeley National Laboratory