Scientists program proteins to pair exactly
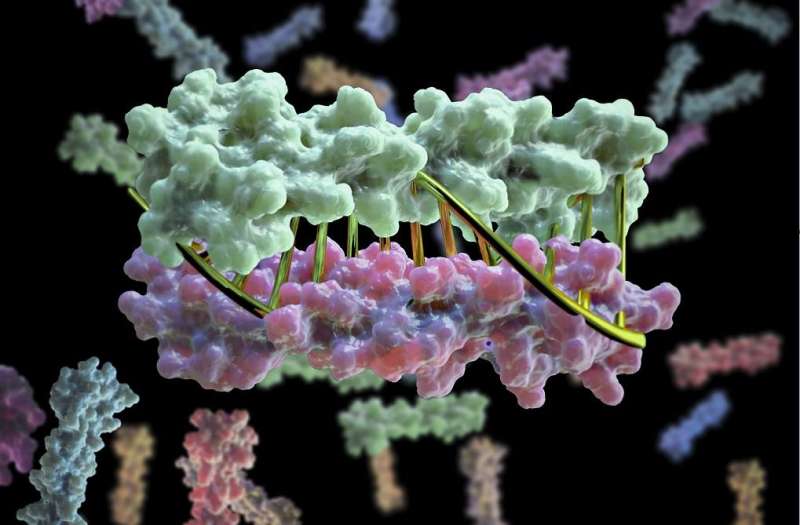
Proteins have now been designed in the lab to zip together in much the same way that DNA molecules zip up to form a double helix. The technique, whose development was led by University of Washington School of Medicine scientists, could enable the design of protein nanomachines that can potentially help diagnose and treat disease, allow for the more exact engineering of cells and perform a wide variety of other tasks.
"For any machine to work, its parts must come together precisely," said Zibo Chen, the lead author of the paper and a UW graduate student in biochemistry. "This technique makes it possible for you to design proteins so they come together exactly how you want them to."
The research was performed at UW Medicine's Institute of Protein Design, directed by David Baker, professor of biochemistry at the University of Washington School of Medicine and a Howard Hughes Medical Institute investigator. The researchers report their findings in the Dec. 19 issue of the journal Nature.
In the past, researchers interested in designing biomolecular nanomachines have often used DNA as a major component. This is because DNA strands come together and form hydrogen bonds to create DNA's double helix, but only if their sequences are complementary.
The team developed new protein design algorithms that produce complementary proteins that precisely pair with each other using the same chemical language of DNA.
"This is a first-of-its-kind breakthrough," Chen said. "What we're doing is computationally designing these hydrogen-bond networks so that each protein pair has a unique complementary sequence. There is only one way for them to come together and they do not cross-react with proteins from other pairs."
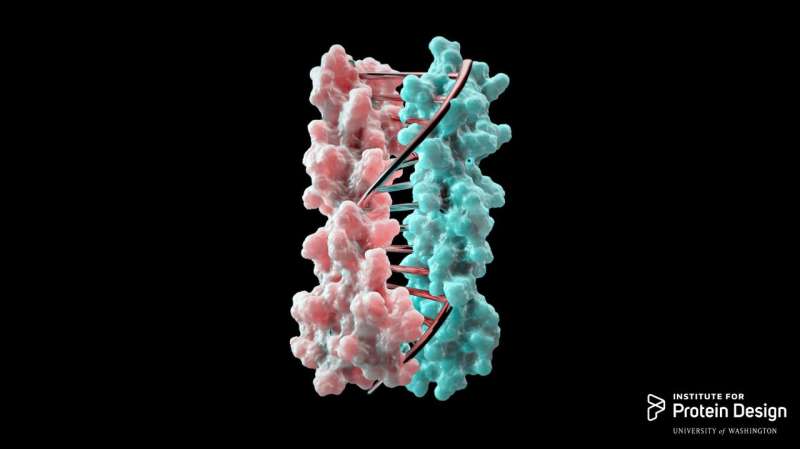
"Engineering cells to do new tasks is the future of medicine and biotechnology, whether that's engineering bacteria to make energy or clean up toxic waste or creating immune cells that attack cancers," said Scott Boyken, another author of the paper and postdoctoral researcher at the Institute for Protein Design. "This technique provides scientists a precise, programmable way to control how protein machines interact, a key step towards achieving these new tasks. We have opened a major door to protein nanomaterial design."
In their study, researchers used a computer program developed in the Baker lab called Rosetta. The program takes advantage of the fact that the shape an amino acid chain will assume is driven by the forces of attraction and repulsion between the amino acids of the chain and the fluid in which the chain is immersed. By calculating the shape that best balances out these forces so that the chain achieves its lowest overall energy level, the program can predict the shape a given amino acid chain will likely take.
More information: Programmable design of orthogonal protein heterodimers, Nature (2018). DOI: 10.1038/s41586-018-0802-y , www.nature.com/articles/s41586-018-0802-y
Journal information: Nature
Provided by University of Washington