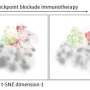
A machine learning approach helps sort and label cell clusters in multiple dimensions

The sorting and automated labelling of cell clusters may be boosted by an algorithm developed by A*STAR researchers. The algorithm facilitates data analysis from a technique, known as cytometry, that effectively sorts and labels cells for use in research.
Cytometry data analysis typically relies on a single-cell process that separates irrelevant cells from populations of interest, according to a set of well-defined parameters. In this selection process, cells are successively divided into subsets according to whether fluorescent markers are present or absent. As the number of parameters to be measured increases, so does the complexity of the process, causing subsets to multiply exponentially. Methods such as dimensionality reduction and cluster analysis have been designed for high dimensions, but often fail to clearly describe the distinguishing characteristics within single cells, which is problematic for new and poorly defined subsets.
To address this, Etienne Becht, Evan Newell, and colleagues from the A*STAR Singapore Immunology Network have created an algorithm, called Hypergate, that generates a strategy that can segregate cells and describe the resulting clusters concisely and accurately. "Similar to solving the Rubik's cube, the algorithm comprises an iterative process in which individual parameter thresholds are adjusted until a composite representing the average of purity and yield is maximized," says Newell.
Hypergate optimizes the size of a high-dimensional rectangle to best encapsulate the target cell cluster. It first includes all cells in the rectangle and modifies the boundaries through successive contraction and expansion phases. Contractions exclude cells to increase purity but can also reduce yield, while expansions increase yield, but sometimes at the expense of purity. "Solving this problem by testing all possible threshold values would be far too computationally expensive and unmanageable with current computers," says Newell.
According to Newell, the resulting gating strategies can provide major insight into the true significance of each cell population. The researchers found that these strategies gave different cell populations from those obtained by traditional gating approaches. They evaluated the sorting ability of Hypergate using innate lymphoid cells, a family of immune cells whose number of subsets is unknown. The algorithm identified two cell clusters with higher purity and yield than existing approaches. It also labelled 24 cell clusters in agreement with previous descriptions, and often with enhanced precision.
Newell's team believes that Hypergate can facilitate flow cytometry and other high-dimensional cell profiling methods, especially to isolate hard-to-define cell populations. "In the future, this could be accomplished in real time by flow cytometry," he says.
More information: Etienne Becht et al. Reverse-engineering flow-cytometry gating strategies for phenotypic labelling and high-performance cell sorting, Bioinformatics (2018). DOI: 10.1093/bioinformatics/bty491
Journal information: Bioinformatics