Tricking bacteria into hydroxylating benzene
Nagoya University researchers use E.coli to convert benzene into phenol, simplifying a chemical reaction that is difficult by conventional methods
Breaking carbon–hydrogen bonds is notoriously difficult in lab chemistry, yet nature does it effortlessly. Now, scientists have used E.coli bacteria to oxidize the C–H bonds in benzene to generate phenol, with a genetically inserted enzyme (cytochrome P450BM3), that originally evolved to target other molecules, long-chain fatty acids.
Getting enzymes to do novel reactions – effectively hijacking the biochemistry of living cells for our own purposes – is generally tricky, involving harsh conditions or genetic modification of the enzymes themselves.
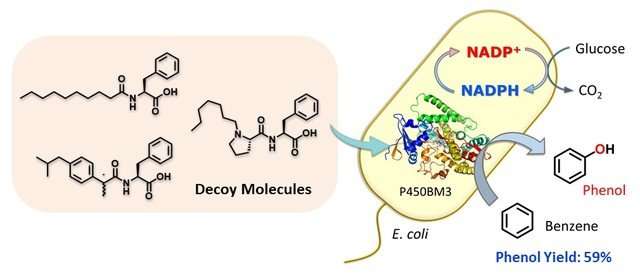
However, researchers at Nagoya University worked around this by using "decoy" molecules, which mimic the native targets (substrates) of naturally occurring enzymes, to activate the desired reaction.
As reported in Angewandte Chemie International Edition, the research team created a compound – dubbed C7-Pro-Phe – based on amino acids. This decoy molecule resembles the fatty acids that E.coli metabolizes. Insert the decoy into an E.coli cell, and it will be mis-recognized as a fatty acid, triggering activation of the inserted P450 enzyme. Now supply benzene (C6H6), and the bacteria get busy, oxidizing C6H6 to C6H6O (phenol). No need for the usual lab kit – living cells can do complex chemistry quietly and efficiently.
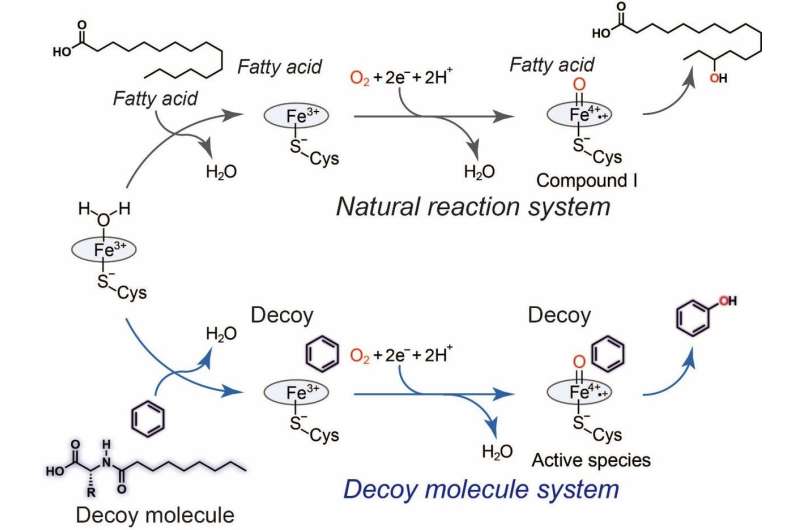
"The advantage of our system is that C7-Pro-Phe can be easily taken up by the bacteria, where it activates P450BM3 in the cell. This effectively turns each bacterium into a whole-cell biocatalyst," study first author Masayuki Karasawa says. "The cell is an optimal setting for the biochemical reaction. The decoys actually remodel the enzyme's active site, giving us control over aspects of the reaction, such as stereoselectivity."
All that is needed is a ready supply of glucose – which can be recycled from waste products – to feed the E.coli.
Because a naturally occurring – rather than genetically modified – variant of the enzyme is expressed by the E.coli, it is likely that other bacteria could also be modified with the same gene to perform this job. Moreover, different decoys might be suitable for different substrates or bacteria. "A combined program of decoy-screening and mutagenesis could create a versatile toolkit for whole-cell reactions using bacteria," co-author Osami Shoji says.
More information: Masayuki Karasawa et al. Whole-Cell Biotransformation of Benzene to Phenol Catalysed by Intracellular Cytochrome P450BM3 Activated by External Additives, Angewandte Chemie International Edition (2018). DOI: 10.1002/anie.201804924
Journal information: Angewandte Chemie International Edition
Provided by Nagoya University