Biophysicists discover how small populations of bacteria survive treatment
Small populations of pathogenic bacteria may be harder to kill off than larger populations because they respond differently to antibiotics, a new study by Emory University finds.
The journal eLife published the research, showing that a population of bacteria containing 100 cells or less responds to antibiotics randomly—not homogeneously like a larger population.
"We've shown that there may be nothing special about bacterial cells that aren't killed by drug therapy—they survive by random chance," says lead author Minsu Kim, an assistant professor in the Department of Physics and a member of Emory's Antibiotic Resistance Center.
"This randomness is a double-edged sword," Kim adds. "On the surface, it makes it more difficult to predict a treatment outcome. But we found a way to manipulate this inherent randomness in a way that clears a small population of bacteria with 100 percent probability. By tuning the growth and death rate of bacteria cells, you can clear small populations of even antibiotic-resistant bacteria using low antibiotic concentrations."
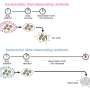
The researchers developed a treatment model using a cocktail of two different classes of antibiotic drugs. They first demonstrated the effectiveness of the model in laboratory experiments on a small population of E. coli bacteria without antibiotic-drug resistance. In later experiments, they found that the model also worked on a small population of clinically-isolated antibiotic-resistant E. coli.
"We hope that our model can help in the development of more sophisticated antibiotic drug protocols—making them more effective at lower doses for some infections," Kim says. "It's important because if you treat a bacterial infection and fail to kill it entirely, that can contribute to antibiotic resistance."
Antibiotic resistance is projected to lead to 300 million premature deaths annually and a global healthcare burden of $100 trillion by 2050, according to the 2014 Review on Antimicrobial Resistance. The epidemic is partly driven by the inability to reliably eradicate infections of antibiotic-susceptible bacteria.
For decades, it was thought that simply reducing the population size of the bacteria to a few hundred cells would be sufficient because the immune system of an infected person can clear out the remaining bacteria.
"More recently, it became clear that small populations of bacteria really matter in the course of an infection," Kim says. "The infectious dose—the number of bacterial cells needed to initiate an infection—turned out to be a few or tens of cells for some species of bacteria and, for others, as low as one cell."
It was not well understood, however, why treatment of bacteria with antibiotics sometimes worked and sometimes failed. Contributing factors may include variations in the immune responses of infected people and possible mutations of bacterial cells to become more virulent.
Kim suspected that something more fundamental was a factor. Research has shown unexpected treatment failure for antibiotic-susceptible infections even in a simple organism like the C. elegans worm, a common model for the study of bacterial virulence.
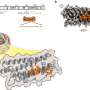
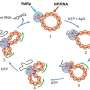
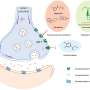
By focusing on small bacteria populations, the Emory team discovered how the dynamics were different from large ones. Antibiotics induce the concentrations of bacterial cells to fluctuate. When the growth rate topped the death rate by random chance, clearance of the bacteria failed.
The researchers used this knowledge to develop a low-dose cocktail drug therapy of two different kinds of antibiotics. They combined a bactericide (which kills bacteria) and a bacteriostat (which slows the growth of bacteria) to manipulate the random fluctuation in the number of cells and boost the probability of the cell death rate topping the growth rate.
Not all antibiotics fit the model and more research is needed to refine the method for applications in a clinical setting.
"We showed that the successful treatment of a bacterial infection with antibiotics is even more complicated than we thought," Kim says. "We hope this knowledge leads to new strategies to fight against infections caused by antibiotic-resistant bacteria."
More information: Jessica Coates et al. Antibiotic-induced population fluctuations and stochastic clearance of bacteria, eLife (2018). DOI: 10.7554/eLife.32976
Journal information: eLife
Provided by Emory University