This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
High speed atomic force microscopy studies provide insights into influenza A viral replication
Researchers at Nano Life Science Institute (WPI-NanoLSI), Kanazawa University, IMDEA Nanoscience (Madrid, Spain) and CNB-CSIC (Madrid, Spain) report in ACS Nano experiments that reveal a cycle of conformational stages that recombinant Influenza A genomes pass through during RNA synthesis.
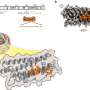
Influenza A is a global health risk responsible for local epidemics and deadly pandemics. As such, the mechanism by which the virus replicates itself has attracted significant interest. Researchers led by Shingo Fukuda at Nano Life Science Institute (WPI-NanoLSI) at Kanazawa University in Japan, Jaime Martin-Benito at CNB-CSIC and Borja Ibarra at IMDEA Nanociencia in Spain, used high-speed atomic force microscopy and electron microscopy to pin down the conformational dynamics of recombinant viral genomes (or rRNPs) during RNA synthesis.
Previous attempts to understand what possible conformational changes occur during Influenza A viral multiplication cycle had been hindered by what the researchers describe as the "bulky double-helical structure" of the viral RNPs (vRNP), which made it hard to see what was going on.
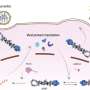
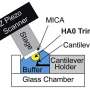
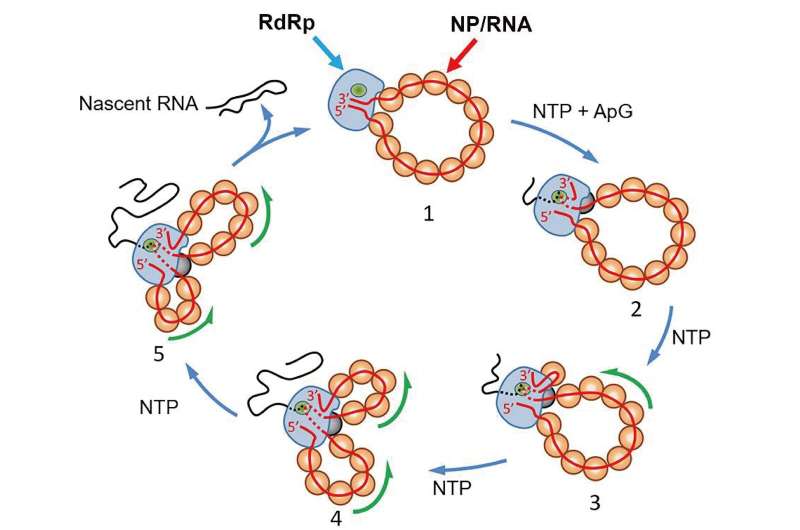
As a result, the researchers produced a circular recombinant ribonucleoprotein complex (rRNP), which allowed them to overcome the "bulky issue." The authors used HS-AFM to follow conformational changes of individual rRNP complexes in real-time during active RNA synthesis.
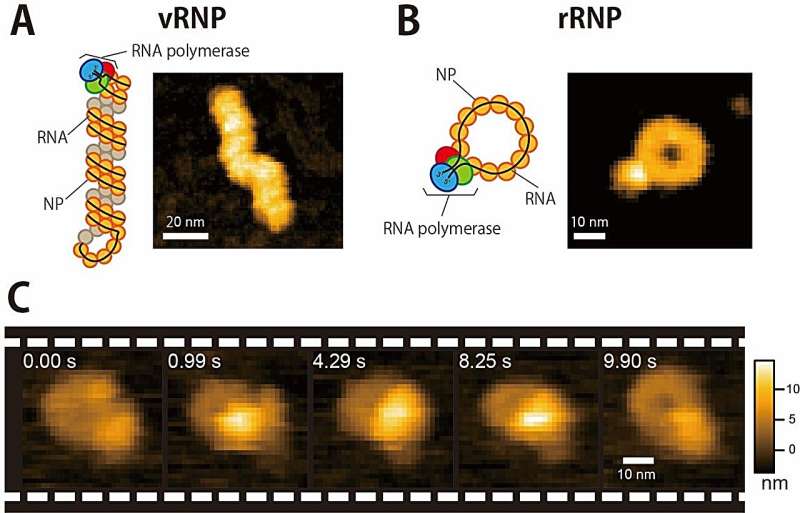
Their work provides the first direct experimental evidence showing that individual rRNPs can be recycled for multiple transcription and replication cycles. This is a key feature for viral multiplication. In addition, their study highlights how factors that affect the stability of the secondary structures of the nascent RNA affect the rate of RNA synthesis.
The authors concluded that the approach is useful for investigating viral transcription and replication mechanisms. "Transcriptional pausing is an intrinsic property of most RNA polymerases, and its regulation constitutes one of the central mechanisms of control of gene expression," they add.
"Future single-molecule experiments of real-time RNA synthesis kinetics by the IAV RdRp [influenza A virus RNA dependent RNA polymerase] within the context of the RNP will help to elucidate the nature of putative pause states and their roles in viral transcription and replication."
More information: Diego Carlero et al, Conformational Dynamics of Influenza A Virus Ribonucleoprotein Complexes during RNA Synthesis, ACS Nano (2024). DOI: 10.1021/acsnano.4c01362
Journal information: ACS Nano
Provided by Kanazawa University