Chemist develops new theory for explaining the function of proteins
A University of Arkansas chemist and his collaborator at North Carolina State University have developed a new theory for explaining how proteins and other biomolecules function based on movement and change of shape and structure rather than content.
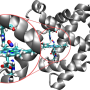
Proteins are considered the workhorse molecules of cells. They are responsible for nearly all tasks in cellular life, including product manufacture, waste cleanup and routine maintenance. For example, some proteins are responsible for transport of materials and information between the cell and its environment, a vital task for the survival and normal function of the cell. Any disorder in protein function could result in disease, and the study of protein function is necessary for understanding the molecular basis of disease.
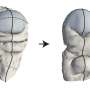
"To function, proteins change their shape," said Mahmoud Moradi, assistant professor of chemistry and biochemistry in the J. William Fulbright College of Arts and Sciences. "Because proteins are not static objects, understanding their conformational dynamics is a necessary step in deciphering the molecular mechanisms underlying their function. The study of protein dynamics is therefore important for understanding the molecular basis of the disease and establishing a 'rational design' for developing more efficient drugs."
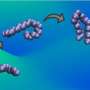
The theory developed by Moradi and Ashkan Fakharzadeh, a graduate student North Carolina State University, describes and simulates the way proteins and other biomolecules change their shape to function.
"Conventional theories of protein dynamics ignore the curved nature of the configurational space of biomolecules," Moradi said. "In this work, we have developed an innovative formalism that relies a geometric theory, traditionally used in general relativity and similar fields, to modify theories of protein dynamics."
Moradi and Fakharzadeh will address two interrelated questions to further develop their theory: How do proteins function by changing their conformation and by undergoing concerted motions, and how can these conformational changes be simulated at an atomic level? Answering these questions would shed light on the structure-function relationships in proteins, Moradi said, and could improve scientists' understanding of diseases at a molecular level.
The researchers' findings were published in the December issue of The Journal of Physical Chemistry Letters, which reports new and original experimental and theoretical research in physical chemistry. A criterion for acceptance in the journal is that the research "reports a significant scientific advance and/or physical insight such that rapid publication is essential."
More information: Ashkan Fakharzadeh et al. Effective Riemannian Diffusion Model for Conformational Dynamics of Biomolecular Systems, The Journal of Physical Chemistry Letters (2016). DOI: 10.1021/acs.jpclett.6b02208
Journal information: Journal of Physical Chemistry Letters
Provided by University of Arkansas