Research on wood formation sheds light on plant biology
(Phys.org)—Scientists at North Carolina State University have discovered a phenomenon never seen before in plants while studying molecular changes inside tree cells as wood is formed.
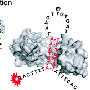
In research published online in Proceedings of the National Academy of Sciences the week of Aug. 20, the team found that one member of a family of proteins called transcription factors took control of a cascade of genes involved in forming wood, which includes a substance called lignin that binds fibers together and gives wood its strength.
The controller protein regulated gene expression on multiple levels, preventing abnormal or stunted plant growth. And it did so in a novel way.
The controller, a spliced variant of the SND1 family, was found in the cytoplasm outside the cell nucleus. This is abnormal, because transcription factor proteins are always in the nucleus. But when one of the four other proteins in its family group was present, the spliced variant was carried into the nucleus, where it bound to the family member, creating a new type of molecule that suppressed the expression of a cascade of genes.
"This is nothing that's been observed before in plants," says Dr. Vincent Chiang, co-director of NC State's Forest Biotechnology Group with Dr. Ron Sederoff. Chiang's research team was the first to produce a transgenic tree with reduced lignin. High lignin levels are desirable for lumber, but lignin is removed during the process of making paper or manufacturing biofuels.
Chiang, a professor in the College of Natural Resources, described the team's finding as the long-sought path to understanding the hierarchy of gene regulation for wood formation.
Abstract: Secondary Wall-Associated NAC Domain 1s (SND1s) are transcription factors (TFs) known to activate a cascade of TF and pathway genes affecting secondary cell wall biosynthesis (xylogenesis) in Arabidopsis and poplars. Elevated SND1 transcriptional activation leads to ectopic xylogenesis and stunted growth. Nothing is known about the upstream regulators of SND1. Here we report the discovery of a stem-differentiating xylem (SDX)-specific alternative SND1 splice variant, PtrSND1-A2IR, that acts as a dominant negative of SND1 transcriptional network genes in Populus trichocarpa. PtrSND1-A2IR derives from PtrSND1-A2, one of the four fully spliced PtrSND1 gene family members (PtrSND1-A1, -A2, -B1, and -B2). Each full-size PtrSND1 activates its own gene, and all four full-size members activate a common MYB gene (PtrMYB021). PtrSND1-A2IR represses the expression of its PtrSND1 member genes and PtrMYB021. Repression of the autoregulation of a TF family member by its only splice variant has not previously been reported in plants. PtrSND1-A2IRlacks DNA binding and transactivation abilities but retains dimerization capability. PtrSND1-A2IR is localized exclusively in cytoplasmic foci. In the presence of any full-size PtrSND1 member, PtrSND1-A2IR is translocated into the nucleus exclusively as a heterodimeric partner with full-size PtrSND1s. Our findings are consistent with a model in which the translocated PtrSND1-A2IR lacking DNA-binding and transactivating abilities can disrupt the function of full-size PtrSND1s, making them nonproductive through heterodimerization, and thereby modulating the SND1 transcriptional network. PtrSND1-A2IR may contribute to transcriptional homeostasis to avoid deleterious effects on xylogenesis and plant growth.
Journal information: Proceedings of the National Academy of Sciences
Provided by North Carolina State University