This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Gene expression technology set to semi-automation
The Human Genome Project generated the first sequence of the human genome, revealing a kind of blueprint of human biology. Two decades later, the field of gene regulatory networks describes a complex system where thousands of genes regulate one another to create appropriate gene expression dynamics.
However, more work is needed to fully understand these networks and determine their precise nature. Current approaches either estimate gene regulation indirectly or utilize accurate and direct but time-consuming, repetitive methods.
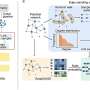
A research group led by Kyoto University has now developed a highly accurate method to semi-automatically estimate gene regulatory networks in multicellular organisms. The method involves measuring time-series gene expression and applying the team's proprietary RENGE computational model. The study is published in the journal Communications Biology.
"RENGE may help identify the key factors for cell differentiation and potentially control the fate of specific cells," says corresponding author Masato Ishikawa of KyotoU's Institute for Life and Medical Science.
Researchers can determine regulatory gene expression using the relatively accurate gene knockout method, where if a gene A is eliminated, a gene B—that A had regulated—is exposed.
The latest single-cell CRISPR technology, which can measure expression changes from large-scale individual knockouts, comes with a caveat: The distinction between directly and indirectly regulated genes is unclear. Knocking out one gene inevitably affects the expression of genes downwind in the network.
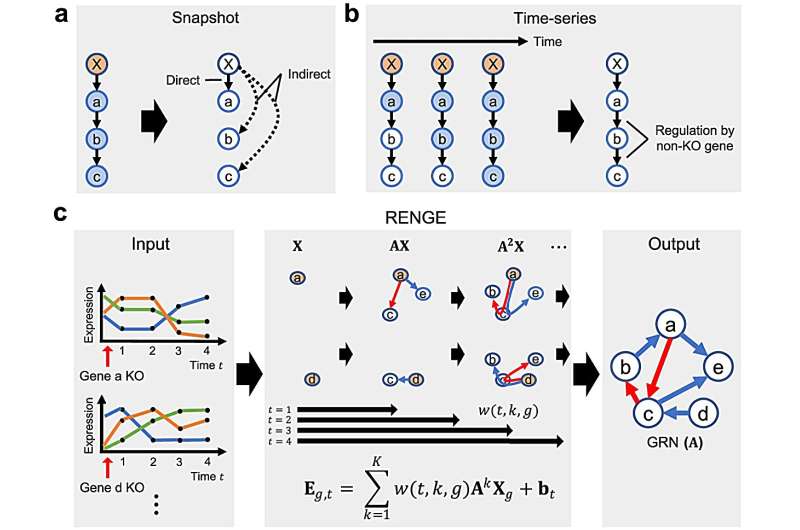
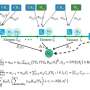
Ishikawa and his colleagues addressed the issue by measuring the expression levels in a time series and adapting their mathematical model to estimate gene regulatory networks.
"When we verified RENGE's accuracy, we obtained results that outperformed existing methods in both simulation data and measured expression data of human iPS cells," adds Ishikawa.
Using RENGE to maximize information obtained from gene knockout, the team could identify a gene regulatory network containing 103 genes expressing human pluripotency, which was highly consistent with findings accumulated over decades of research in molecular biology.
"However, the beauty is that we could obtain the same results from only one RENGE experiment. Furthermore, we found that transcription factors that regulate the same target genes in this regulatory network tend to work as protein complexes, among which one may be a key factor for pluripotency," elaborates Ishikawa.
The inherent versatility of this method allows for estimating regulatory networks in various other life systems beyond iPS cells.
"RENGE may inspire new technologies such as producing artificially manipulated cells with specific functions," remarks Ishikawa.
More information: Masato Ishikawa et al, RENGE infers gene regulatory networks using time-series single-cell RNA-seq data with CRISPR perturbations, Communications Biology (2023). DOI: 10.1038/s42003-023-05594-4
Provided by Kyoto University