This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Research provides insight into constructing gene regulatory networks
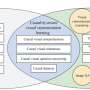
Gene regulatory networks (GRNs) depict the regulatory mechanisms of genes within cellular systems as a network, offering vital insights for understanding cell processes and molecular interactions that determine cellular phenotypes. Transcriptional regulation, a prevalent type for regulating gene expression, involves the control of target genes (TGs) by transcription factors (TFs).
One of the major challenges in inferring GRNs is to establish causal relationships, rather than just correlation, among the various components of the system. Therefore, inferring gene regulatory networks from the perspective of causality is essential for understanding the underlying mechanisms that govern the dynamics of cellular systems.
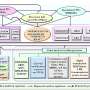
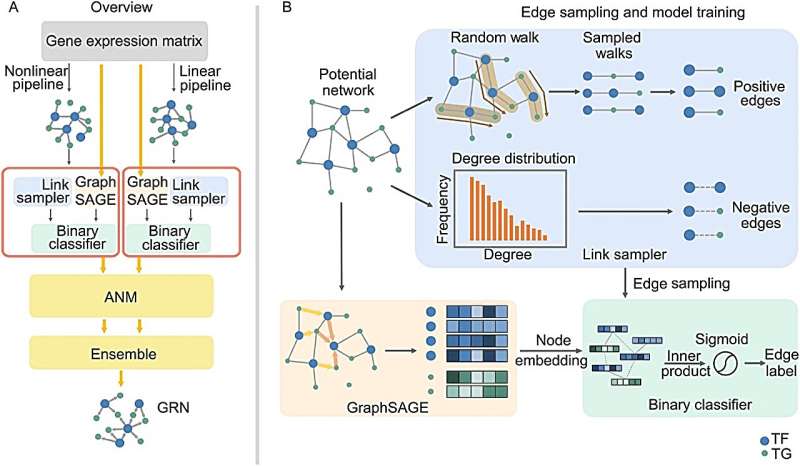
Quantitative Biology has published an approach, titled "Gene Regulatory Network Inference based on Causal Discovery Integrating with Graph Neural Network," that leverages graph representation learning and causal asymmetric learning while taking into account both linear and non-linear regulatory relationships. GRINCD achieves superior performance in predicting the regulatory relationships of not only TF-TG but also TF-TF, where generalized correlation-based methods are unattainable.
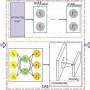
GRINCD applies ensemble learning to predict the causal regulation of each regulator-target pair based on additive noise model (ANM) which takes high-quality representation for each gene generated by Graph Neural Network as input.
Specifically, GRINCD utilizes random walk and nodes' degree distribution to generate edge labels and feeds them to a two-layer GraphSAGE connected with a binary classifier for obtaining the representation of each node.
GRINCD achieves optimal performance on multiple datasets under various evaluation metrics. As an application, through analyzing the substantial alterations in regulatory relationships with disease progression, GRINCD identifies crucial potential regulators that drive the transition from colon inflammation to colon cancer.
More information: Ke Feng et al, Gene regulatory network inference based on causal discovery integrating with graph neural network, Quantitative Biology (2023). DOI: 10.1002/qub2.26
Provided by Higher Education Press