This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Deciphering the fragrance code: High-quality sequencing and analysis of the 'XiangQingCai' (XQC) genome
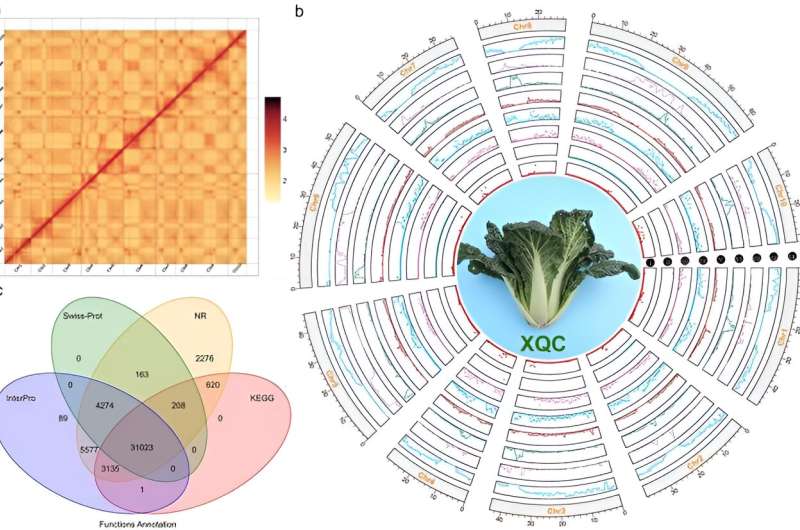
'Vanilla' (XQC, brassica variety chinensis), a Brassica rapa subspecies chinensis, is a vital vegetable crop in the Brassicaceae family, renowned for its intense volatile fragrance. Despite the significant progress that has been made in understanding the genomes of Brassica species, including the discovery of a whole-genome triplication event, XQC's genome remains unexplored.
In September 2023, Horticulture Research published research titled "The high-quality sequencing of the Brassica rapa 'XiangQingCai' genome and exploration of genome evolution and genes related to volatile aroma."
In this study, the genome of XiangQingCai (XQC) was sequenced utilizing a combination of PacBio HiFi, Illumina, and Hi-C technologies, providing a detailed genomic map to explore fragrance synthesis in Brassica. The assembly process yielded 150.73 Gb of genomic data, resulting in a high-quality assembled genome of approximately 466.10 Mb with notable contig and scaffold N50 sizes, demonstrating the assembly's accuracy and completeness.
This extensive dataset facilitated the mapping of 92.47% of sequences to 10 XQC chromosomes and achieved a read mapping rate exceeding 99.75%, underscoring the genome's comprehensive coverage. Genome annotation revealed a significant proportion (59.50%) of the genome as repetitive sequences, with a detailed gene prediction identifying 47,570 genes, with 99.57% being annotated against major databases.
The study analyzed gene families in XQC and related species, revealing 49,906 gene families, with specific attention to gene families unique to XQC. Divergence time estimations indicated a close relationship and synteny with B. rapa QGC and Pakchoi, pinpointing a divergence time frame of 1.1–1.4 million years ago.
The exploration of polyploidization events through Ks value analysis indicated the occurrence of four significant polyploidization events, including a recent whole-genome triplication. Syntenic and global alignment analyses provided insights into gene loss and retention post-polyploidization, enhancing our understanding of XQC's genomic evolution.
Furthermore, the study identified genes involved in the terpenoid biosynthesis pathway, crucial for volatile aroma formation, and highlighted the TPS gene family's evolution, which plays a vital role in terpenoid biosynthesis.
In summary, this analysis not only establishes the chromosomal-level genome of XQC but also lays the groundwork for future genomic studies of B. rapa, offering rich resources for understanding the molecular mechanisms behind fragrance synthesis and the regulatory networks within Brassicaceae.
More information: Zhaokun Liu et al, The high-quality sequencing of the Brassica rapa 'XiangQingCai' genome and exploration of genome evolution and genes related to volatile aroma, Horticulture Research (2023). DOI: 10.1093/hr/uhad187
Provided by NanJing Agricultural University




















