This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Unveiling the high-quality MicroTom genome and its impact on developmental biology
Tomato (Solanum lycopersicum) has marked its global significance with annual production of approximately 186 million tons in 2020. In addition to its edible and economic value, tomato has become a vital model in developmental biology research, surpassing Arabidopsis thaliana in some cases like fruit development, metabolism, plant-pathogen interactions, and symbiosis studies.
MicroTom is a tomato cultivar and currently a widely applied experimental model plant for laboratory studies due to its smaller size, shorter growth cycle and higher transformation efficiency.
Despite the extensive re-sequencing of numerous tomato cultivars, a high-quality genome for MicroTom has been conspicuously absent, presenting a challenge for developmental biologists who rely on the Heinz tomato genome for reference but use MicroTom for functional experiments.
In July 2023, Horticulture Research published a research article titled "Comprehensive regulatory networks for tomato organ development based on the genome and RNAome of MicroTom tomato." In this study, researchers provided a high-quality genome of MicroTom and conducted comparative genomic analysis with the previously published Heinz tomato genome.
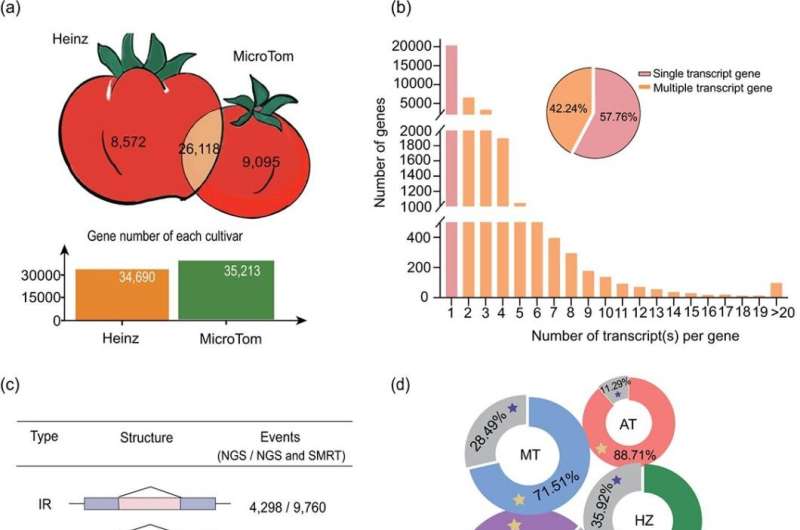
First, they obtained a MicroTom genome assembly of 799 Mb with 60 contigs using a combination of Nanopore and Illumina sequencing data. Protein-coding gene annotation of the MicroTom assembly captures 98.57% of the Embryophyta BUSCO (odb10) genes.
Comparative genomics revealed substantial similarities and differences between the MicroTom and Heinz genomes, with both sharing common polyploidy events but displaying considerable genomic divergence in terms of gene content, structural variations, and single-nucleotide polymorphisms (SNPs).
Additionally, researchers present the RNAome landscape of MicroTom across different organ/developmental stages/treatments and performed comprehensive analyses of the transcriptome of MicroTom protein-coding genes, with respect to gene expression and alternative splicing (AS).
Then, they constructed multiple gene co-expression networks with the reference genome and abundant gene expression data, which will provide valuable clues for the identification of important genes involved in diverse regulatory pathways during plant growth, e.g. arbuscular mycorrhizal symbiosis and fruit development.
The study also shed light on non-coding RNAs, including miRNAs, lncRNAs, and circRNAs, by integrating data from different developmental stages and conditions.
A total of 210 miRNAs were identified from the sRNA datasets, of which 164 belong to 48 known tomato miRNA families in miRbase. A total of 4,835 lncRNAs were annotated from MicroTom transcripts, and >2,944 of them (60.89%) were transcribed from intergenic regions. And 19,840 circRNAs supported by three independent software tools were identified by mapping the sequencing reads onto the MicroTom genome.
Finally, researchers developed a comprehensive database (MicroTomBase), providing online search and download possibilities for their data and results. This online resource is designed to be a valuable tool for researchers working with the MicroTom tomato, enhancing studies in comparative genomics, gene expression, and functional genomics.
In summary, this study not only presents a high-quality MicroTom genome but also offers a rich annotation of both coding and non-coding elements, backed by extensive transcriptomic analyses.
The insights into gene expression profiles, alternative splicing, non-coding RNA functions, and co-expression networks significantly advance our understanding of the genomic and transcriptomic complexity in the MicroTom tomato, offering a valuable resource for future research in plant biology and genetics.
More information: Jia-Yu Xue et al, Comprehensive regulatory networks for tomato organ development based on the genome and RNAome of MicroTom tomato, Horticulture Research (2023). DOI: 10.1093/hr/uhad147
Journal information: Horticulture Research
Provided by Plant Phenomics