New version of Chinese tree shrew genome annotation released

A research team led by Prof. Yao Yonggang from Kunming Institute of Zoology of the Chinese Academy of Sciences has reported a new version of Chinese tree shrew genome annotation, and studied multiple genomic and transcriptomic characteristics of Chinese tree shrew, including gene family expansion, transcriptomic similarity across four species, and host immune response against virus infection. The study was published in Zoological Research.
Prof. Yao's team has assembled the first high-quality genome of the Chinese tree shrew (KIZ version 1: TS_1.0) using high depth (~79X) short-read sequencing technology and the first chromosome-level tree shrew genome (KIZ version 2: TS_2.0) using single-molecule real-time (SMRT) sequencing technology. In this study, the team improved the genome annotation of Chinese tree shrew by using transcriptome sequencing data of various origin.
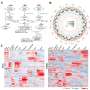
The researchers collected high-quality RNA-seq datasets of tree shrews from publicly available sources, as well as two ISO-seq datasets and 139 RNA-seq datasets newly generated in this study. They obtained a total of 53,298 newly annotated coding transcripts and 115,562 newly annotated non-coding transcripts, and produced a relatively complete and reliable tree shrew genome annotation (KIZ version 3:TS_3.0).
Based on this comprehensive annotation, the researchers explored the spatial expression and alternative splicing patterns of the tree shrew transcripts and characterized the orthologous relationships among tree shrews and other species. They also compared expression similarity across species and found that the species clustering patterns based on expression data from brain, liver, testis, kidney, and heart tissues showed distant divergence of mice from primates and tree shrews.
Besides, the researchers identified 13 pathways including neuro-related pathways such as Parkinson's disease and Alzheimer's disease between tree shrews and humans which showed greater protein sequence identity than those between mice and humans. They profiled the expression patterns of five pathways related to the brain and showed that tree shrews are closer to primates than to mice at the transcriptomic level.
In addition, the researchers profiled the transcriptome patterns of host immune responses to viral infection using the TS_3.0 genome annotation, and found that newly annotated genes such as purine rich element binding protein A and tree shrew specific expansion STT3 oligosaccharyltransferase complex catalytic subunit B gene family were dysregulated upon virus infection, which may suggest their important role during host immune response.
The improved annotation of tree shrew genome illuminated the unique and common genetic features of tree shrews.
More information: Mao-Sen Ye et al, Comprehensive annotation of the Chinese tree shrew genome by large-scale RNA sequencing and long-read isoform sequencing, Zoological Research (2021). DOI: 10.24272/j.issn.2095-8137.2021.272
Provided by Chinese Academy of Sciences