This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Researchers describe new technological platform to accelerate drug development
Drug development is currently an extremely long, expensive and inefficient process. Findings generated in a lab are often very hard to replicate once translated into animal models or in humans.
A family of pharmacological targets, on which approximately 35% of FDA-approved drugs work, consists of receptors at the surface of cells named "G protein coupled receptors" or GPCRs. These membrane proteins sense a wide variety of signals from outside the cell, from hormones to neurotransmitters, or light and odors, to trigger responses of cells, from immune reactions to muscle contraction.
In a new study, researchers at Boston University Chobanian & Avedisian School of Medicine describe the development of a new technological platform that will enhance the accuracy and efficiency of drug discovery related to a large family of important pharmacological targets, i.e., GPCRs, allowing it to obtain information that was previously unattainable.
The work appears in Cell.
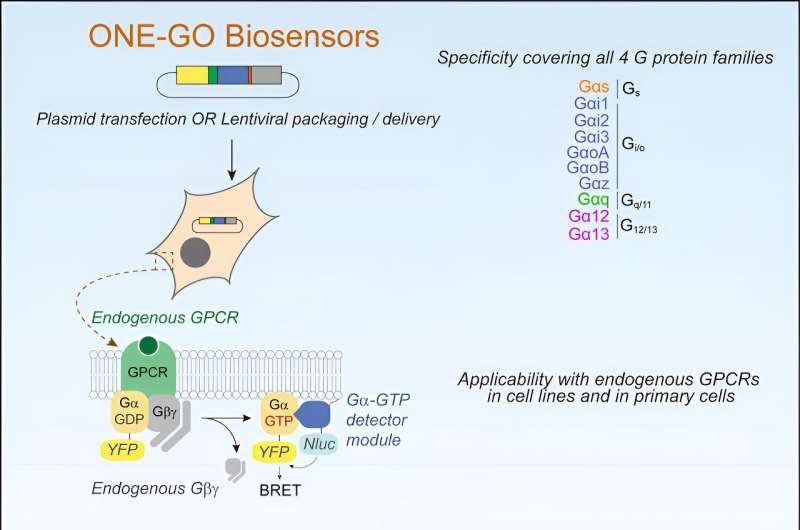
"The new technological platform [consists of] biosensors that allow us to detect the activity of GPCRs in their native cellular context and in real time not only with unprecedented fidelity, but also with relative ease. A novelty of this platform is that it is easy for others to implement and that it has been made readily available through a public repository such as open-source," explains Remi Janicot, first author of the study and a Ph.D. student in the laboratory of Mikel Garcia-Marcos, Ph.D., professor of biochemistry and cell biology at the school.
The researchers used molecular engineering to create these biosensors, artificial genes that—once expressed as proteins in cells—could detect the activation of very specific cellular responses in real time. The researchers then showed that these biosensors could be leveraged to characterize how many different natural ligands and synthetic drugs change cell responses when the act on the large GPCR class of receptors.
According to the researchers, the biosensor platform described could accelerate drug development for any disease in which GPCRs are implicated, which is a virtually endless list covering neurodegenerative disorders, cancers, cardiovascular diseases, obesity, and asthma, to name a few.
"For example, the ongoing interest in developing safer opioid or non-opioid analgesics would benefit from the features of the technology described in our work. Similarly, there is tremendous interest in developing orally bioavailable substitutes for the anti-obesity drugs commercialized as Ozempic and Wegovy, which would also benefit from the technology we described," said Garcia-Marcos.
The researchers hope the tools developed in this study will transform how the field of pharmacology and drug development approaches the investigation of a large and important class of targets.
"Since medicines acting on these targets are already part of everyday life, we see great potential in accelerating the discovery of improved drugs for many different indications, from analgesia to anti-obesity drugs," added Janicot.
More information: Remi Janicot et al, Direct interrogation of context-dependent GPCR activity with a universal biosensor platform, Cell (2024). DOI: 10.1016/j.cell.2024.01.028
Provided by Boston University School of Medicine