This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Study shows 3D organization of DNA controls cell identity programs
In a complex called chromatin, long strands of DNA in cells' nuclei are tightly wrapped around a scaffolding of proteins, like a rolled-up ball of yarn. A new study by Weill Cornell Medicine investigators reveals that beyond providing a convenient way to store DNA in a tight space, the 3D organization of noncoding gene regulators in chromatin contributes to the control of key cell identity programs in early embryonic development.
The results have implications for understanding this critical period and how changes in the 3D chromatin architecture may contribute to developmental abnormalities or diseases such as diabetes or cancer.
The study, published in Nature Structural & Molecular Biology, provides new insights into a complex layer of genetic regulation. It shows that at the earliest phases of embryonic development, changes in the 3D organization of DNA dictate cell fate decisions. Noncoding stretches of DNA, called enhancers, which regulate the spatiotemporal expression of other genes, play a pivotal role.
How the DNA is folded in the tiny nucleus brings these enhancers close to genes they regulate and dictates which gene expression programs are turned on. They show that this type of 3D epigenetic regulation is critical for committing to one of the first cell lineages formed in an embryo. They also identify highly interacting "3D hubs," clusters of enhancers and genes, that are vital in dictating these cell fates.
"If you want to understand the regulation of gene expression or gene expression programs, it is absolutely critical to understand the three-dimensional organization of the DNA," said lead author Effie Apostolou, associate professor of molecular biology in medicine at Weill Cornell Medicine. "It is also essential to understanding how 3D DNA misfolding may result in dysregulated gene programs in disease."
Not long ago, scientists referred to noncoding stretches of DNA as "junk DNA," Apostolou said. However, scientists have learned that noncoding stretches of DNA play a critical role in controlling the expression of other genes, often while residing at large distance from the genes, and that mutating, silencing or rewiring these enhancers can cause disease. But with hundreds of thousands of potential noncoding enhancers in the genome, matching them with the genes they control and predicting which ones are impactful remains very tricky.
"It has been very difficult to identify functional enhancers and assign them to the correct target gene or genes and determine whether enhancers work together in a synergistic, competing, or redundant way," she said. "We now believe many of these challenges can be addressed by understanding how all these elements interact in the 3D nucleus."
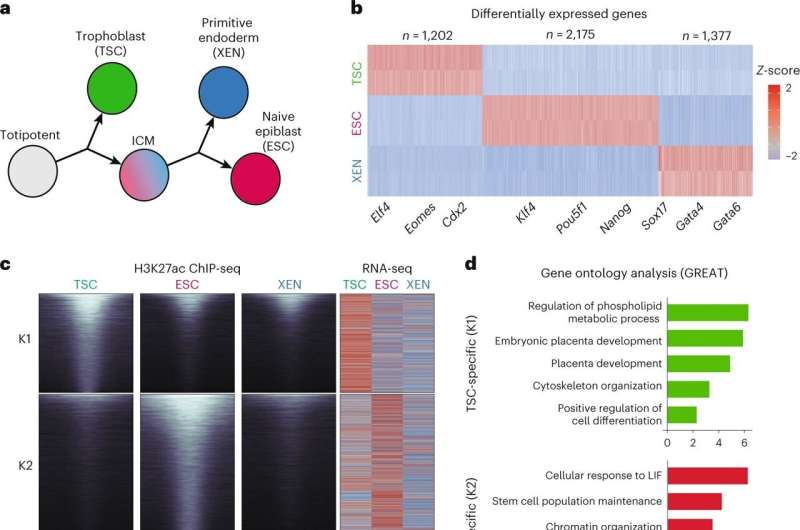
In the study, Apostolou and colleagues examined the 3D architecture of folded DNA in the nucleus during the earliest phases of embryonic development in an animal model. They show that as the cells begin to change into the first three distinct cell lineages, there is marked remodeling in the nucleus that results in variations in the 3D architecture of the DNA among the cell types.
These differences dictate gene expression patterns in each cell type. They found the most active and cell-type specific (or signature) genes clustered in "3D hubs" in the respective cell type.
"We mapped the 3D organization of DNA in the first three cell lines during early development and used the maps to find critical noncoding gene enhancers that help control the fate of each cell line," she said.
Study co-author Alexander Polyzos, a computational instructor of molecular biology in medicine at Weill Cornell Medicine, built a machine learning model of these 3D interactions to predict which genes will be turned on—and how much—in each cell type during early development. The model was more accurate than previous models that did not consider this level of 3D organization.
"By mapping 3D interactions, we got a much better understanding of what controls gene expression and how genes can coordinately change their levels during the transition between different cell fates," said Apostolou, who is also a member of the Sandra and Edward Meyer Cancer Center at Weill Cornell Medicine.
The 3D organization of the DNA in a particular cell type may bring the most important genes and gene enhancers in that cell type physically close to each other to enable their interactions, she explained. By contrast, the investigators also showed that the activity of "housekeeping genes," which are essential to basic cell functions and active regardless of cell type, are less affected by the 3D organization and interactions with distal enhancers.
Then, they used the model to systematically test what happens when you "break" this organization by targeting individual enhancers or hubs. They found that targeting hubs had the greatest impact on gene expression.
The next steps for the team are to study 3D packaging of DNA and gene expression using human cells. They are collaborating with other investigators to determine how disruption of this 3D architecture may contribute to diseases like cancer or diabetes.
"We want to build more models to understand how enhancers and genes communicate in the 3D nucleus and what changes in the context of different diseases," Apostolou said.
The research also adds another layer to scientists' understanding of epigenetic regulation of cell identity. Apostolou explained that initially, scientists focused on identifying coding genes and what they do. Next, they studied noncoding genetic elements that turn genes on and off. Now, they must also consider the 3D organization of both.
"Understanding the 3D architecture of DNA packaging brings all of this together to understand better genes, their regulators, and how they interact and uncover new ways to modulate these complex relationships to reprogram cell identity," she said.
More information: Dylan Murphy et al, 3D Enhancer–promoter networks provide predictive features for gene expression and coregulation in early embryonic lineages, Nature Structural & Molecular Biology (2023). DOI: 10.1038/s41594-023-01130-4
Journal information: Nature Structural & Molecular Biology
Provided by Cornell University