This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Ribosomal protein exhibits remarkable evolutionary transformation
A team of researchers from the Universities of Newcastle and Umeå has discovered that a ribosomal protein exhibits a remarkable evolutionary transformation, with its three-dimensional structure changing drastically while its sequence remains relatively conserved.
The protein, known as msL1/msL2, is found in ribosomes of parasitic microorganisms called microsporidia, and it is suggested to play a role in stabilizing the highly reduced protein synthesis machinery in these unique organisms. The study is published in the journal Molecular Biology and Evolution.
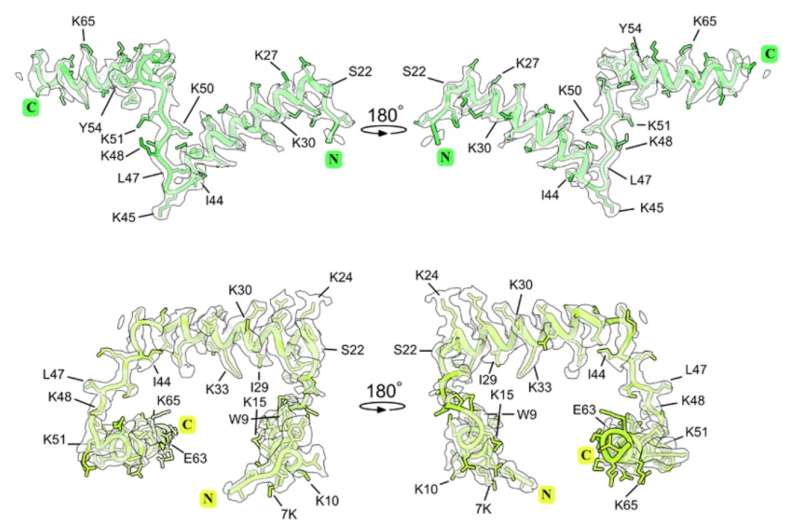
"Despite its conserved sequence, msL1/msL2 adopts distinct folds in two different microsporidian species, Encephalitozoon cuniculi, and Vairimorpha necatrix. This structural divergence is particularly striking given that the two proteins share approximately 41% sequence similarity," says Léon Schierholz, one of the authors from Umeå University.
The different folds of msL1/msL2 result from changes in their binding sites within the ribosome. In E. cuniculi ribosomes, msL2-Ec binds to a shorter helix of ribosomal RNA (rRNA) than its counterpart in V. necatrix ribosomes, msL1-Vn. This difference in rRNA sequence drives the proteins to adopt different folds to maintain their interactions with the ribosome.
The researchers also found that the different folds of msL1/msL2 are not accompanied by any loss of function. Both proteins can perform their role in binding to and stabilizing the ribosome, suggesting that the protein's structure is not tightly linked to its function in this case.
This study provides a new perspective on the evolution of ribosomal proteins, particularly those with intrinsically disordered regions. It suggests that the structure of these proteins can evolve more rapidly than their sequence and that this evolution is driven by changes in their binding partners within the ribosome.
"Evolution is hard to study because we often fail to see the intermediate stages, or 'transition forms,' between two distinct organisms or molecules in living bodies. Instead, we tend to observe abrupt and seemingly radical changes. In this study, we believe we have identified one of these rare transition forms, providing a deeper insight into how biological molecules change as they evolve," says Sergey Melnikov, last author of the study, from Newcastle University School of Medicine.
The findings of this study have important implications for our understanding of protein evolution and the factors that shape protein structure and function. They also highlight the challenges of using sequence similarity to predict protein structure and function, particularly for intrinsically disordered proteins.
More information: Léon Schierholz et al, A conserved ribosomal protein has entirely dissimilar structures in different organisms, Molecular Biology and Evolution (2023). DOI: 10.1093/molbev/msad254
Journal information: Molecular Biology and Evolution
Provided by Umea University