This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Reassessing what we can expect from peptides in disease detection
Based on blood tests, it is possible to detect rare genetic diseases, recognize cancer, or determine the inflammation level in the body. Moreover, due to the rapid development of medical diagnostics based on biofluid analysis, many efforts are being made worldwide to adapt medical approaches, making personalized medicine the paradigm of future health care.
Moving in this direction, scientists at the Institute of Physical Chemistry of the Polish Academy of Sciences (IPC PAS) presented their research on unique peptides for use in novel sensors for the rapid and simple detection of many diseases.
Inflammation is the body's natural defense mechanism against pathogens or harmful chemicals causing acute symptoms such as pain, swelling, redness, or bruising. This mechanism involves the body's intense production of specific molecules, such as cytokines, which are used against infection, and inflammation can be easily diagnosed with such symptoms.
However, some inflammatory reactions are not visible and can last much longer than a sudden infection. If inflammatory cells remain for a long time, chronic inflammation can develop. Chronic inflammation can indicate a variety of health problems, including autoimmune diseases and cancers, which are difficult to overcome.
Detection of the inflammation process in the body can be easily performed from the blood, while the monitoring of the inflammation course is a completely different story. Usually, the infection rate is evaluated by focusing on the level of C-reactive protein known as CRP, a common early-rising biomarker for various inflammatory conditions in the body.
Its appearance in plasm is based on the body's response to the increased cytokines level during inflammation when the liver produces CRP. Its level can also distinguish inflammation cause; for example, the increase in CRP level can relate to viral infections, while very high is characteristic of bacterial infections.
The blood levels of this ring-shaped molecule change rapidly and pronouncedly with any inflammation defense not only for infections but also as a response to tissue damage that also triggers inflammation in all types of tissues. So far, the monitoring of CRP levels can deliver valuable and precise information about the disease progression.
Following this field, a researcher from the IPC PAS, Dr. Katarzyna Szot-Karpińska, studied the protein-peptide interactions, particularly between CRP and CRP-binding materials. She focused on peptides due to their higher stability under harsh conditions to degradation, lower cost, and the possibility of more efficient use in biomedical sensors than antibodies, which are the most common CRP receptors.
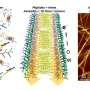
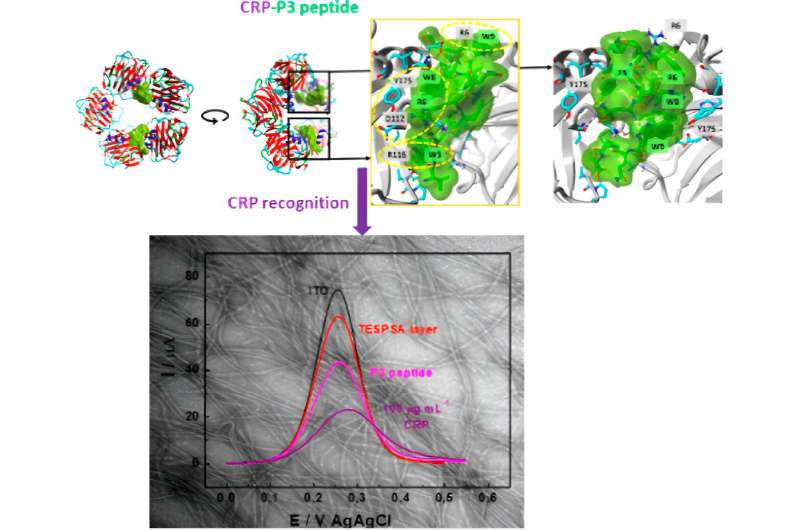
In a paper published in the journal Analytical Chemistry, the researchers used a phage-display method to identify CRP-binding phages. Identifying such phages enables the determination of the sequence of peptides exposed on the surface of the phages with a high affinity towards CRP.
A key driver of the study was the issue of monitoring long-term inflammation using an electrochemical platform modified with stable molecules as an alternative to antibodies. The selected peptides were synthesized and fully characterized, and their interactions with CRP were studied, resulting in the identification of the three most promising phage-derived CRP-binding molecules.
The peptide with the highest affinity towards CRP was immobilized on electrodes for later use as an electrochemical CRP sensor. Biological and physicochemical techniques were used to understand the mechanisms of protein-peptide and protein-peptide interactions.
Dr. Katarzyna Szot-Karpińska, a scientist working on this project, said, "In our studies, for the first time, we demonstrated that a single 12-mer peptide identified from a phage library has been used for CRP recognition. The identified peptides were characterized, and their interactions with CRP were studied for application in a sensing platform showing the new approach to biomedical sensor development."
The results were surprising, showing a new field for electrochemical research. The new material on the electrode showed up to two orders of magnitude higher affinity for CRP than the antibodies used in ELISA. In addition, the detection efficiency was promising for the selected peptide, even in the presence of three interfering proteins. Moreover, no additional chemicals were needed, making the detection more environmentally friendly than with classical techniques.
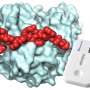
Additionally, the theoretical simulations, including computational modeling analyses performed in collaboration with Prof. Sławomir Filipek from the University of Warsaw, were implemented to complement the biological and physicochemical studies and delivered startling results.
In silico studies explained the specifics of the binding of particular peptides to CRP. The results show which peptide from a list of options is the best CRP binder and confirm the experimental study. So far, combining experimental and theoretical studies optimized the screening process, thereby accelerating research.
"The added value of this work is the integration of experimental methods with computational modeling analysis. The modeling from known amino acid sequences of peptides confirms that the P3 peptide is the best binder for CRP. Such a combined approach has not been reported previously and demonstrates how numerical methods/in silico analysis can replace or enhance laborious experimental techniques."
"Using molecular docking to identify the best binders eliminates the application of chemicals, which is vital to developing greener chemistry. This study validates the numerical approach to identifying peptide binding properties and represents an important step on the road to peptide-based sensors. Moreover, if we know how the docking works, in the future, we could tailor the sequence of the peptide by changing, for example, one of the amino acids to obtain the best binding molecule to the studied target, like disease biomarkers," says Dr. Szot Karpińska
Such a computational approach to CRP sensing using peptides has never been described before, and it illustrates a new trend in research in which computations can support arduous experimental techniques.
The proposed method enables monitoring of the inflammation course, making it possible to check the CRP level selectively and sensitively using much smaller and more stable molecules than are currently used. This work would not be possible without engaging many different methods and techniques from various fields, proving the deep need for interdisciplinarity in science, especially in health protection.
It shows the importance of combining experimental studies with an in-depth study of intermolecular interactions. The described research can be a game-changer in inflammation diagnostics and treatment, especially in the case of long-term inflammation courses or even novel lab-on-the-chip devices for personalized medicine, drug development, and drug delivery. The team is looking for new receptors for disease markers and new solutions for studying molecular interactions.
More information: Katarzyna Szot-Karpińska et al, Investigation of Peptides for Molecular Recognition of C-Reactive Protein–Theoretical and Experimental Studies, Analytical Chemistry (2023). DOI: 10.1021/acs.analchem.3c03127
Journal information: Analytical Chemistry
Provided by Polish Academy of Sciences