This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Korean study develops efficient SNP marker system for lettuce cultivar identification
Lettuce (L. sativa L., 2n = 2x = 18) is an agriculturally important leafy vegetable cultivated predominantly in temperate climates and produced in large quantities in Korea. The complete decoding of its genome and the analysis of around 45,000 genes have advanced cultivar development. However, distinguishing closely related lettuce cultivars remains a challenge, particularly due to their morphological diversity and similar genetic profiles.
Traditional methods, such as the distinctness, uniformity, and stability (DUS) test based on morphological features, are labor-intensive and subject to environmental conditions. Therefore, the application and improvement of molecular markers such as SSR and SNP for genetic analysis is necessary for more accurate and efficient lettuce cultivar identification and genetic studies.
In May 2022, Horticulture Research published a research article titled "Genome-wide core sets of SNP markers and Fluidigm assays for rapid and effective genotypic identification of Korean cultivars of lettuce (Lactuca sativa L.)."
In this study, researchers developed and validated core sets of genome-wide single nucleotide polymorphism (SNP) markers for lettuce cultivar identification using genotyping-by-sequencing (GBS) and SNP-genotyping approaches, followed by high-throughput analysis on the Fluidigm platform.
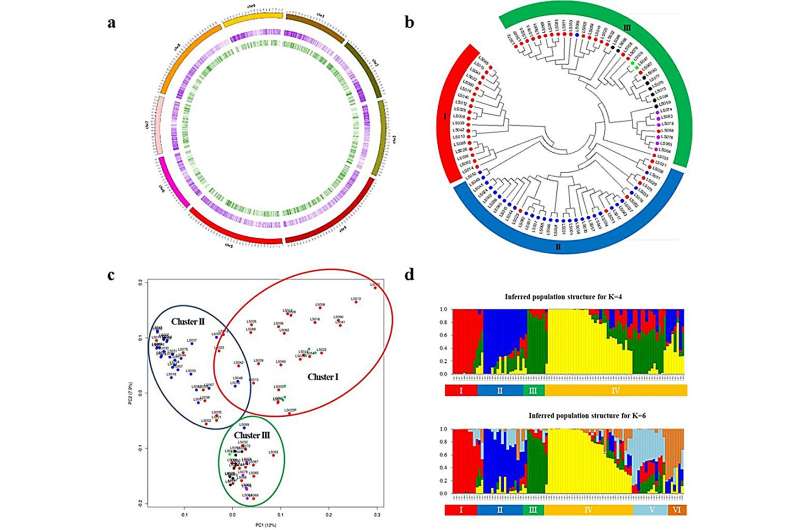
GBS identified 17,877 high-quality SNPs for 90 commercial lettuce cultivars. Using these SNPs, phylogenetic analysis classified the 90 cultivars into three distinct groups, largely aligning with their horticultural types. Principal component analysis (PCA) and population structure analysis using these SNPs yielded similar clustering results, thus reinforcing the close association between SNP genotypes and horticultural types.
To develop effective SNP markers for cultivar identification, 294 SNPs with significant polymorphism were selected. The core set of 192, 96, 48 and 24 markers was further screened and validated using the Fluidigm platform. Phylogenetic analysis based on the core set of all SNPs successfully differentiated the cultivars identified so far.
These core SNP marker sets will be useful for constructing a lettuce DNA database for cultivar identification and purity testing, as well as for supporting DUS (Distinctness, Uniformity, and Stability) examination under the UPOV-based Plant Variety Protection (PVP) system. Finally, the study validated the 24 core markers with Fluidigm genotyping on DNA mixtures, demonstrating their ability to identify both homozygous and heterozygous lines.
In summary, this study successfully developed and validated core sets of SNP markers for lettuce cultivar identification through genotyping by sequencing (GBS). This approach is expected to significantly improve the efficiency and accuracy of lettuce cultivar identification, providing a valuable tool for future genetic research and practical agricultural applications.
More information: Jee-Soo Park et al, Genome-wide core sets of SNP markers and Fluidigm assays for rapid and effective genotypic identification of Korean cultivars of lettuce (Lactuca sativa L.), Horticulture Research (2022). DOI: 10.1093/hr/uhac119
Journal information: Horticulture Research
Provided by NanJing Agricultural University