This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Study shows how AI can detect antibiotic resistance in as little as 30 minutes
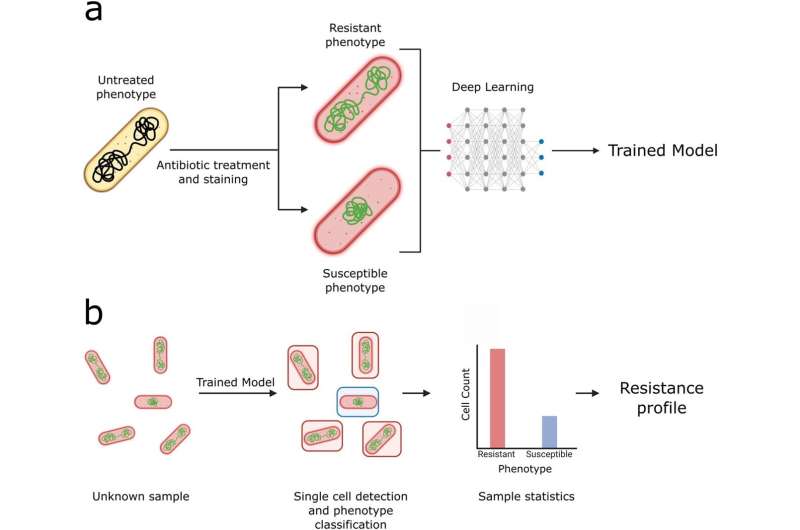
To mark World Antimicrobial Awareness Week, researchers supported by the Oxford Martin Program on Antimicrobial Resistance Testing at the University of Oxford have reported advances towards a novel and rapid antimicrobial susceptibility test that can return results within as little as 30 minutes—significantly faster than current gold-standard approaches.
The study, "Deep learning and single-cell phenotyping for rapid antimicrobial susceptibility detection in Escherichia coli," has been published in Communications Biology.
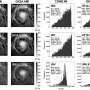
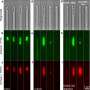
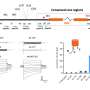
In their study, the team used a combination of fluorescence microscopy and artificial intelligence (AI) to detect antimicrobial resistance (AMR). This method relies on training deep-learning models to analyze bacterial cell images and detect structural changes that may occur in cells when they are treated with antibiotics. The method was shown to be effective across multiple antibiotics, achieving at least 80% accuracy on a per-cell basis.
The researchers say their model could be used to identify whether cells in clinical samples are resistant to a range of a wide variety of antibiotics in the future.
Co-author of the paper Achillefs Kapanidis, Professor of Biological Physics and Director of the Oxford Martin Program on Antimicrobial Resistance Testing, said, "Antibiotics that stop the growth of bacterial cells also change how cells look under a microscope, and affect cellular structures such as the bacterial chromosome. Our AI-based approach detects such changes reliably and rapidly. Equally, if a cell is resistant, the changes we selected are absent, and this forms the basis for detecting antibiotic resistance."
The researchers tested their method on a range of clinical isolates of E. coli, each with varying levels of resistance to the antibiotic ciprofloxacin. The deep-learning models were able to detect antibiotic resistance reliably and at least 10 times faster than established state-of-the art clinical methods considered to be gold standard.
The team hopes to continue developing their method so that it becomes faster and more scalable for clinical use, as well as adapting its usage for different types of bacteria and antibiotics.
According to the Global Research on Antimicrobial Resistance (GRAM) Project—a partnership involving the University—almost 1.3 million people died in 2019 due to AMR.
Current testing methods rely on growing bacterial colonies in the presence of antibiotics. However, such tests are slow, often requiring several days to understand how resistant bacteria are to a range of antibiotics.
This can be problematic when patients have potentially life-threatening infections, such as sepsis, requiring urgent treatment. This usually forces doctors to either prescribe specific antibiotics based on their clinical experience or a cocktail of antibiotics known to be effective across multiple bacterial infections.
However, if ineffective antibiotics are prescribed the patients' infections may get worse and they will need to be treated with more antibiotics. One potential outcome of this is increased AMR to antibiotics in the community.
The researchers say that if developed further, the rapid nature of their method may facilitate targeted antibiotic treatments—helping to decrease treatment times, minimize side effects, and ultimately slow down the rise of AMR.
Co-author of the paper Aleksander Zagajewski, doctoral student with the University's Department of Physics, said, "Time is beginning to run out for our antibiotic arsenal; we are hoping our novel diagnostics will pave the way for a new generation of precision treatments for the most sick patients."
More information: Alexander Zagajewski et al, Deep learning and single-cell phenotyping for rapid antimicrobial susceptibility detection in Escherichia coli, Communications Biology (2023). DOI: 10.1038/s42003-023-05524-4
Journal information: Communications Biology
Provided by University of Oxford