This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Detection and extraction of similar features in disease-related gene groups
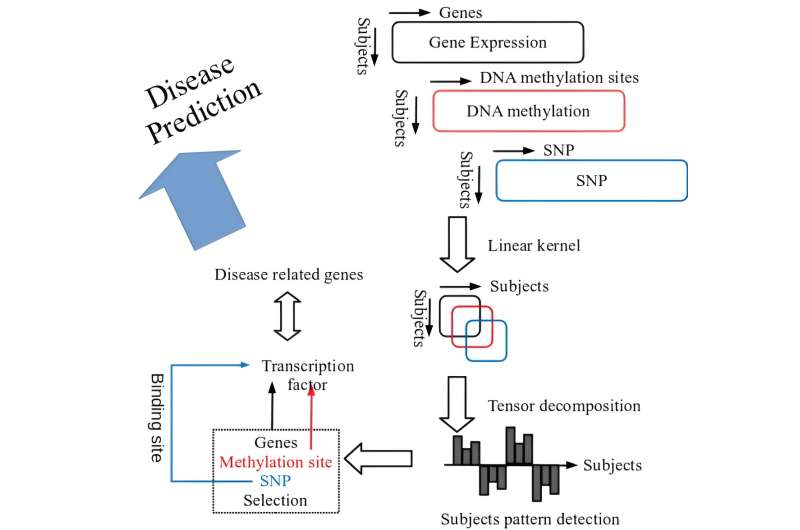
Multiomics analysis that integrates different layers of profiles altogether is challenging, since the number of variables in profile substantially differ from each other. For instance, gene expression profile and genomic DNA methylation profile are often analyzed together; however, there are only tens of thousands of genes, whereas the number of DNA methylation sites are as many as tens of millions.
The numbers differ by orders of magnitude and the number of pairs between gene and DNA methylation sites are enormous. As such, it requires huge computational resources to conduct integrated analysis without controlling target numbers by focusing on DNA methylation sites in specific regions, such as promoter regions, based on prior knowledge. However, limiting the genomic regions to be analyzed, effects of DNA methylation on other regions (e.g., enhancer) and functions remain unexplored.
The study and the accomplishment
Published in the journal PLOS ONE, a recent study applied the method developed in the previous study to treat multiomics data (gene expression profile, DNA methylation profile, Single Nucleotide Polymorphism (SNP) profile) which Iwate Tohoku Medical Megabank Organization (IMM) comprehensively collected from 100 local resident participants, and confirmed whether the relationships with disease-related gene can be identified or not.
This is the data-driven approach called the variable extraction method which employed kernel tensor decomposition-based unsupervised study (hereafter called tensor decomposition), and this method is applicable to the datasets with all subjects belonging to a healthy group.
In addition, this method is implementable with kernel sized (square of the subject participants, in particular) or so memories per 1 profile, and thus, even for the enormous profiles such as genome and epigenome comprising tens of millions of SNP or DNA methylation sites, data-driven analysis can identify unique patterns across study subjects and identify the variables (gene expression profile, DNA methylation profile, SNP profile) that exhibit similarity to those patterns.
In this study, tensor decomposition was applied to multiomics data of each autosome retrieved from three cell types, CD4 positive T cells, monocytes, and neutrophils. As a result, the two patterns of subjects profiles were identified, and these two patterns of subjects observed in 22 autosomes show very strong mutual correlations between the other autosomes. As the genes identified in each autosome are completely independent from one another, it suggests that the observed patterns shared across chromosomes are not coincidence.
The observed orthogonal patterns also cannot be explained by batch effects, and it is improbable that the same batch effects are present in the three omics profiles obtained by the different methodologies. These two patterns of subjects are obtained as the second and the third singular value vectors by tensor decompositions, respectively. The second singular value vector was detected from all three cell types while the third singular value vector was detected from two cell types except monocytes.
Then, the genes and DNA methylation regions with homological profiles with these patterns were selected to find out that these genes and regions are targeted by many transcription factors. Furthermore, the enrichment analysis revealed that these transcription factors relate to various diseases.
In addition, the study found that identified SNP are statistically and significantly overlapped with binding sites of these transcription factors. Therefore, the authors believe that the application of tensor decompositions is effective for the integrated analysis of multiomics datasets.
More information: Y-h. Taguchi et al, Integrated analysis of human DNA methylation, gene expression, and genomic variation in iMETHYL database using kernel tensor decomposition-based unsupervised feature extraction, PLOS ONE (2023). DOI: 10.1371/journal.pone.0289029
Journal information: PLoS ONE
Provided by Chuo University