This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Study finds linkage between DNA methylome and RNA transcriptome in Fe deficiency in tomato roots
Tomato (Solanum lycopersicum), one of the most important horticultural crops, is rich in iron and vitamins. As an essential micro-element for plants, iron (Fe) deficiency not only restricts tomato growth, development and fruit quality, but also poses health risks to animals and humans, resulting in human nutritional disorder worldwide.
While the transcriptional and post-transcriptional mechanisms of plant responses to Fe-deficiency have been extensively studied, the contribution of epigenetic modulations, such as DNA methylation, remains poorly understood. Here, we reported the linkage between DNA methylome and RNA transcriptome in response to both short-term (12 hours) and long-term (72 hours) Fe deficiency in tomato roots.
By combining methylome analysis and genetic approach, we proposed that dynamic epigenetic DNA methylation in the CG context at the bHLH39 promoter is involved in its transcriptional regulation, thus contributing to the Fe deficiency-induced responses of tomato.
The article "CG hypermethylation of the bHLH39 promoter regulates its expression and Fe deficiency responses in tomato roots" has been published in Horticulture Research.
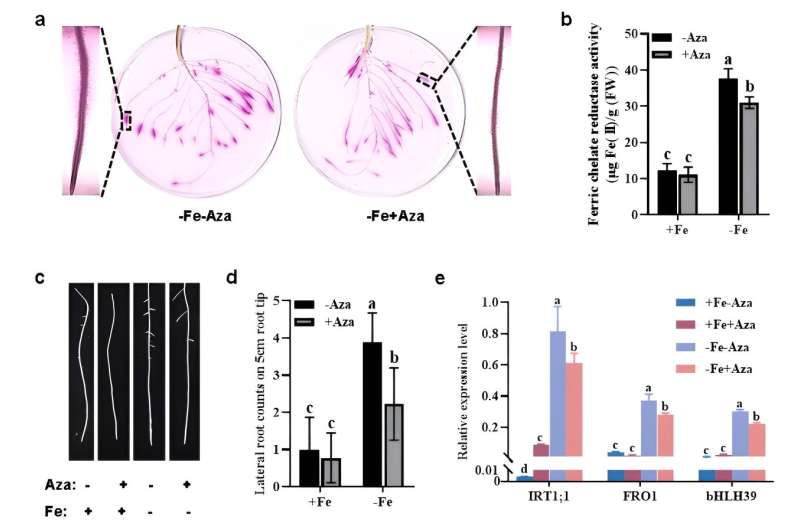
Firstly, 5–azacytidine (Aza), a DNA methylase inhibitor, was exploited to investigate its influence on the expression of Fe deficiency–induced responses in tomato. As shown in Figure 1, exogenous application of Aza greatly repressed the induction of root FCR activity, so did the Fe deficiency–induced development of root morphology and the transcript levels of three core genes (IRT1;1, FRO1 and bHLH39), suggesting the importance of DNA methylation in regulating Fe deficiency responses.
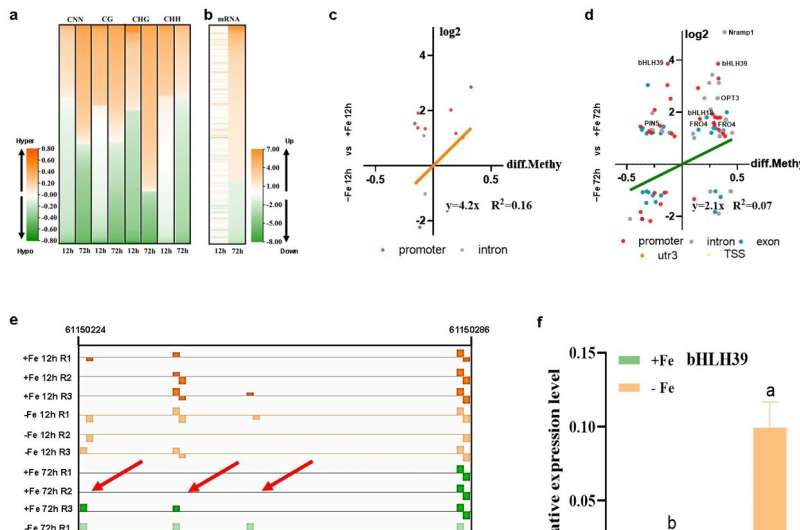
Subsequently, dynamic changes of DNA methylation and gene expression identified that the number of hyper−differentially methylated regions (DMRs) increased over time, especially in the CG and CHG contexts, which was accompanied by greater transcript levels of Fe deficiency–induced genes (Figure 2a, b).
Furthermore, we detected no obvious correlation between the changes in DNA methylation levels and the changes in transcript levels of the affected genes under either short−term or long−term treatments, with the exception of some core Fe deficiency-induced genes, i.e., bHLH39, bHLH18, Nramp1, OPT3 and FRO4, whose expression changes displayed some relationships with DNA methylation changes under long−term treatment (Figure 2c, d).
Notably, more hypermethylation than hypomethylation at CG sites in the bHLH39 promoter, which was associated with the induction of bHLH39 expression by long−term Fe deficiency treatment (Figure 2e, f). In agreement, we detected lower CG methylation at the bHLH39 promoter and lower bHLH39 expression but more severe chlorosis phenotype in MET1−RNA interference lines compared to wild−type seedlings (Figure 2g-i), which further confirmed the positive interconnection between promoter CG methylation and gene transcription.
In summary, we revealed the dynamics of both DNA methylation and the transcriptome, which is important for Fe deficiency–induced responses in tomato roots.
Furthermore, DNA methylation in the CG context is positively related to gene expression and mediates the transcription of specific genes, such as bHLH39, which contributes to Fe deficiency responses in tomato roots. The results will enrich our understanding of regulatory mechanism under Fe deficiency in plants, as well as provided theoretical and technical support for crop improvement.
More information: Huihui Zhu et al, CG hypermethylation of the bHLH39 promoter regulates its expression and Fe deficiency responses in tomato roots, Horticulture Research (2023). DOI: 10.1093/hr/uhad104
Provided by NanJing Agricultural University





















