This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Extreme environments leave genomic imprint in tiny organisms: Study
The signature written in genomic DNA has long been linked to ancestry, not to geographic location. But a recent study using AI from Western University, published in Scientific Reports, provides evidence that living in extreme temperature environments leaves a discernible imprint on the genomes of microbial extremophiles.
Using machine learning, an interdisciplinary research team co-led by Western biology associate professor Kathleen Hill and computer science adjunct professor Lila Kari found that, unexpectedly, the genomic signatures of microbial extremophiles living in comparable extreme environments are similar, even though they belong to two different domains of the Tree of Life, namely Bacteria and Archaea.
"This discovery flies in the face of conventional thinking that pervasive, genome-wide, genomic signatures carry only information about naming, describing and classification of organisms," said Hill, an expert in gene mutation, population genetics and genome evolution.
Extremophiles live in exceedingly harsh environments like volcanoes, deep-sea trenches and polar regions, all characterized by extreme conditions (high temperature, radiation, pressure or acidity), that would pose an existential threat to most other living organisms. For example, Pyrococcus furiosus is an archaeum (single-celled organism) first discovered thriving at 100 C near a volcanic vent in Italy, while Chryseobacterium greenlandensis is a bacterium that survived 120,000 years within the ice of a glacier in Greenland.
"This is similar to someone living in the Arctic finding out their DNA is more similar to algae that grows in the Arctic, than to the DNA of their cousin," said Kari, an expert in biodiversity, data science and machine learning. "DNA should be mostly about inheritance, biological relatedness, and common ancestry, not about the place you live in, but we see something completely different with extremophiles."
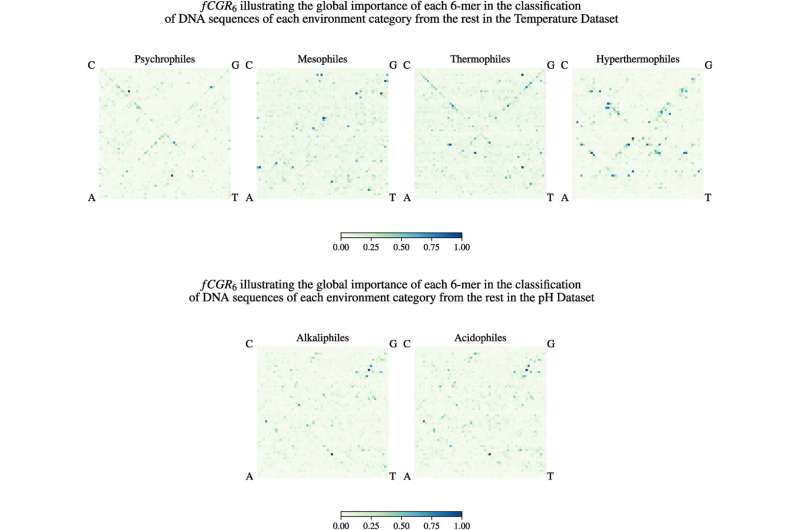
For the study, Kari, Hill and their collaborators used supervised and unsupervised machine learning to analyze genomic signatures. The supervised AI algorithm was trained on DNA sequences with taxonomic labels (bacteria or archaea). It learned to recognize genomic patterns that characterize taxonomy, and it was then able to predict the taxonomy of unknown DNA sequences with high accuracy.
Surprisingly, when trained with the same DNA sequences labeled instead with the type of environment the organisms lived in (hot or very cold), the AI algorithm learned some genomic patterns that are associated with the environment type. Moreover, it could then predict, with medium-high accuracy, which kind of extreme environment an unknown DNA sequence came from.
"We did not expect this outcome and it gave us the idea to continue exploring," said Kari, also a professor at University of Waterloo's School of Computer Science.
Double-checking the results
To double-check these positive results, the team used unsupervised learning with the same dataset as input, only this time around the DNA sequences had no labels at all.
In other words, the DNA sequences were fed to an unsupervised AI algorithm that did not know anything about either their taxonomy or environment, and was asked a simple question: "By looking at these DNA sequences, which ones look more similar to you?"
This blind AI algorithm successfully produced clusters of similar sequences, with each cluster containing sequences with similar genomic patterns. Surprisingly, some of the clusters that formed contained both bacteria and archaea sequences, even though bacteria and archaea are taxonomically less like each other than a bear is to a fungus.
"Upon a closer examination, these unlikely bedfellows in the cluster turned out to inhabit the same extreme environment," said Kari. "This means not only does an extreme environment signal exist in the very fabric of the DNA of extremophiles, but in some cases, this extreme environment signal drowns out the biological relatedness signal."
The identification of an environmental signal in the genomic signature of extremophiles holds remarkable implications, for example, for the future of space exploration.
"By understanding how these resilient organisms adapt to extreme conditions on Earth, we can potentially harness their unique capabilities," said Hill. "This discovery brings us closer to unlocking the secrets of survival in harsh extraterrestrial environments, opening doors to new frontiers, and expanding our possibilities in space."
More information: Pablo Millán Arias et al, Environment and taxonomy shape the genomic signature of prokaryotic extremophiles, Scientific Reports (2023). DOI: 10.1038/s41598-023-42518-y
Journal information: Scientific Reports
Provided by University of Western Ontario