July 5, 2023 feature
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
New study presents whole-body gene expression atlas of an adult metazoan
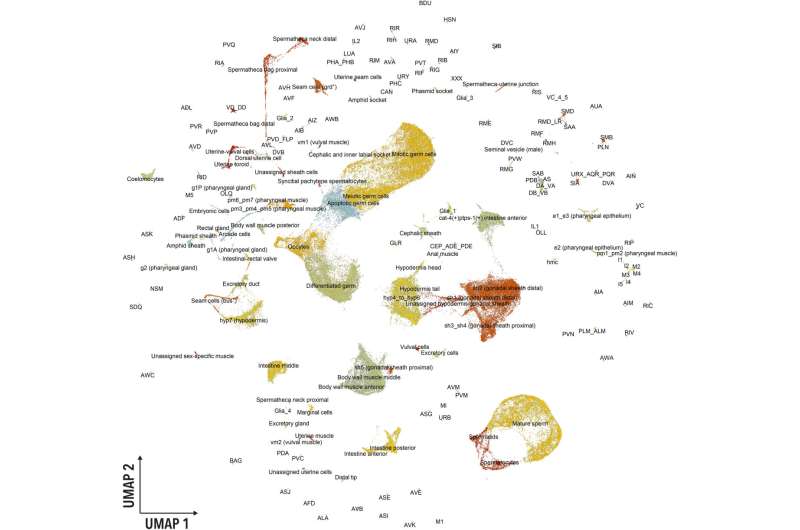
Cellular identity is defined by gene activity that drives intercellular communication and is pivotal to understand mechanisms underlying multicellular organisms. In a new report Abbas Ghaddar and a research team in biology, pediatrics, systems biology, genome sciences, bioengineering and cardiovascular research in the U.S., presented an atlas of gene activity of a fertile adult metazoan Caenorhabditis elegans at the level of single-cell resolution. The compendium comprised 180 distinct cell types and a whopping 19,657 expressed genes. The outcomes are now published in Science Advances.
The team predicted large-scale transcription factor expression profiles associated with defining cellular identity and thousands of intercellular interactions across the organism, alongside ligand-receptor pairs mediating them. The team found a list of genes conserved across phyla with consistent expression across cell types involved in basic and essential functions, and presented them as experimentally validated housekeeping genes. They explored this data by using the Wormseq application to advance current understandings of molecular mechanisms underlying the functional capacity of multicellular organisms and perturbations associated with their malfunctions.
Developing a gene expression atlas at the single-cell level
The growth and reproduction of multicellular organisms are sustained by a diverse range of cell types. Even the more simpler animal types such as C. elegans develop according to a select plan to discern food quality, find mates and escape predators. Typically, the functions of C. elegans are carried out by about 20 broadly defined cell types. Such cell types are regulated through specific gene networks.
Unveiling the molecular mechanisms of functional multicellular organisms requires cataloging gene expression and discerning their identity alongside capacity for intercellular communications. Such outcomes can inform future studies on the impact of genetic, chemical and environmental triggers that alter gene expression at the cellular level.
Recent advances have shown the scope of analyzing single-cell transcriptomics of C. elegans to observe single-cell gene expression profiles at the embryo, larvae and adult stages of the organism. In this work, Ghadder and colleagues used a catalog of adult gene expression to explore several concepts, including:
- The concept of housekeeping genes.
- Transcription factor-mediated cellular identity.
- Molecular drivers of cell-cell interaction.
The researchers developed a website (Wormseq.org) to categorize the data gathered from this study, providing a platform for biologists to manipulate C. elegans and experimentally test several hundreds of hypotheses and predictions presented herein.
Identifying C. elegans cell types and subtypes as single-cell clusters
The research team harvested wild-type hermaphrodite C. elegans as young adults defined by the vulva morphology and the presence of less than five eggs. The team conducted single-cell RNA sequencing to collect three independent biological replicates and used a filter to remove any low quality or damaged cells. After processing the cells following the Monocle 3 pipeline; a powerful software toolkit for single-cell analysis, the results indicated differences in the batches although the replicates accurately represented average cell types and gene expression profiles.
The team annotated the clusters using previously published single-cell RNA sequencing data and the existing rich literature on C. elegans tissue. The team identified gene markers and manually annotated each cluster, to provide a rationale to each annotation. They recognized broad cell types in the adult hermaphrodite including the intestine, neurons, and XXX cells, to name a few.
During the classification, they unexpectedly identified transcriptionally distinct spermatheca subpopulations and noted the presence of cell types typically absent in adult hermaphrodite C. elegans, such as seminal vesicle gene markers exclusive to males.
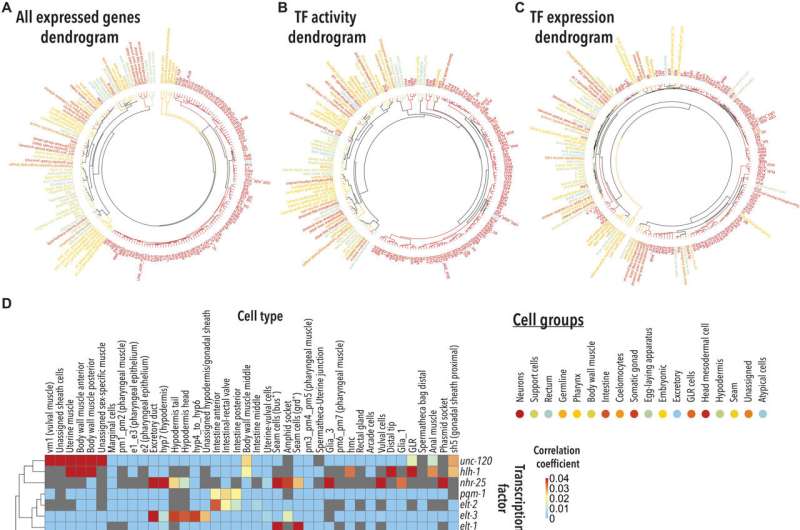
Identifying house-keeping genes
Housekeeping genes provide insight to intriguing biological questions, including which genes underly cellular function in eukaryotes. Housekeeping genes are also an interesting reference across molecular biology and biochemistry assays.
Since the single-cell RNA-sequencing data is yet unavailable for C. elegans, the team subjected the organisms to genetic or chemical triggers to examine the outcomes within a specific timeframe with the animal models. They studied if the housekeeping genes met the common criteria to define their "housekeepingness," namely their consistent expression across cell types and conditions including essentiality and conservation.
Understanding the interactions underlying cellular identity
Ghaddar and the team used the single-cell RNA-sequencing dataset of the C. elegans larval stage to obtain insights to regulatory mechanisms driving cell-specific gene expression and explored transcription factor-mediated gene expression profiles. Based on the predictions obtained therein, they determined cell type associations to explore cellular identity.
They also conducted studies on the relationship between the predicted transcription factor activity and cell identity associations, and cellular functions. The outcomes make it possible to explore a range of hypotheses across several fields of study.
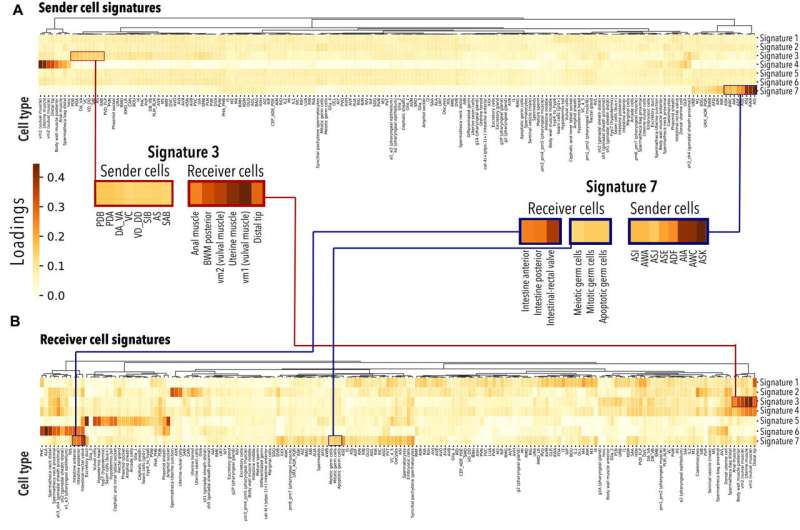
Whole body reconstruction cell-to-cell interactions
The cell-to-cell interactions played a critical role to maintain tissue and organ systems sustaining metazoan life. Using single-cell transcriptomics datasets, the researchers developed a tool Cell2Cell to interpret cell-cell interactions from the expression of ligand- and receptor-encoding genes. Using several existing databases and machine learning methods such as Tensor-cell2cell, the team identified meaningful hypotheses about molecules during cell-to-cell communication and cell-to-functional interactions in C. elegans.
Outlook
In this way, Abbas Ghaddar and colleagues presented a comprehensive single-cell atlas of a wild-type adult C. elegans organism. Since life scientists had previously generated single-cell transcriptional analyses for other metazoans including mice and humans, this single cell RNA-sequencing dataset was defined by specific features of interest. The level of gene activity uncovered during this study helped answer several questions. For example, which of the genes were relevant to maintain cell identity and function? Which genes reflect biological or experimental noise?
The team intend to incorporate additional sing-cell proteomic data to increase the accuracy of functional predictions in the future. The outcomes of this work provide a major stepping stone to identify key molecular players, and the tools developed in this work can facilitate a guide to develop similar predictive technologies to investigate more complex organisms.
More information: Abbas Ghaddar et al, Whole-body gene expression atlas of an adult metazoan, Science Advances (2023). DOI: 10.1126/sciadv.adg0506
W. Clay Spencer et al, Isolation of Specific Neurons from C. elegans Larvae for Gene Expression Profiling, PLoS ONE (2014). DOI: 10.1371/journal.pone.0112102
Journal information: Science Advances , PLoS ONE
© 2023 Science X Network