Mathematical methods for analyzing single-cell transcriptomic data
Single-cell transcriptomics is revolutionizing the field of cell biology. This technique enables researchers to analyze the genes expressed in individual cells, which provides more information on the different cell types within tissues compared to bulk transcriptomics.
Most current analytical methods for single-cell transcriptomic data assume that cell types are discrete. However, cell types are more nuanced. For example, a cell can change type during development, cell types can be subclassified and even cells of the same type may vary in their gene expression patterns. To recognize the less discrete and more continuous nature of gene expression between cell types, more advanced analytical methods are needed for single-cell transcriptomic data.
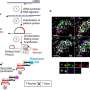
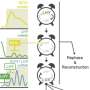
Mathematicians from the laboratory of Ludwig Oxford's Helen Byrne and the University of Oxford's Heather Harrington have teamed up with cell biologists from Xin Lu's group in a joint Ludwig Oxford-Mathematical Institute project. They have developed three methods based on topological data analysis, which describes the shape of the data and the relationships between the components. These methods take into account both discrete and continuous transcriptional patterns equally across multiple scales of resolution simultaneously.
When applied to an existing single-cell transcriptomic dataset, the methods validate the previously observed gene expression patterns. However, they also identify some additional biologically meaningful genes, demonstrating the enhanced analytical power of these mathematical methods. Their work is published in Entropy.
More information: Renee S. Hoekzema et al, Multiscale Methods for Signal Selection in Single-Cell Data, Entropy (2022). DOI: 10.3390/e24081116
Provided by Ludwig Cancer Research