Live-seq: Sequencing a cell without killing it

RNA sequencing allows scientists to study the expression of genes in a cell. Since messenger RNA (mRNA) is generated from a DNA gene, that information can be used to identify the original gene sequence and thus measure the activity of thousands of genes (i.e. the transcriptome) in these cells.
The challenge with RNA sequencing is to capture mRNA from a specific cell out of a mixed population—which is how most cells exist in nature. To do this, scientists have developed "single-cell RNA sequencing" (scRNA-seq), which provides unprecedented insights for basic and biomedical research, and even drug development.
Introducing Live-seq
Now, scientists from the groups of professors Bart Deplancke at EPFL and Julia Vorholt at ETH Zurich have addressed a problem with scRNA-seq: needing to lyse (kill) cells. The researchers, led by Dr. Wanze Chen (EPFL) and Dr. Orane Guillaume Gentil (ETH Zurich), present Live-seq. The innovative approach keeps the cells alive during RNA extraction for further study while also being minimally invasive.
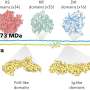
The key to Live-seq is a microscopy technique called "fluidic force microscopy," or FluidFM, which uses microscopic channels—thinner than human hair—to manipulate tiny volumes of fluids (femtoliters) in a sample under the microscope developed at ETH Zurich some years back. Because of this, FluidFM allows users to insert substances into individual cells, or to extract cytoplasm including mRNA from single cells without having to kill them.
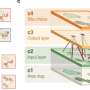
The critical advance leading up to Live-seq emerged when the researchers managed to preserve and read out the mRNA (the transcriptome), from these tiny amounts of cytoplasmic sample. As a result, Live-seq can now connect a cell's transcriptome at a given time to its later molecular or phenotypic behavior, i.e. to monitor the activity of thousands of genes in a single cell at discrete time points—what the scientists refer to as a "temporal" transcriptomic analysis.
"With Live-seq, we can now uniquely address highly interesting and biomedically relevant questions, such as why certain cells differentiate and sister cells do not, or why certain cells are resistant to a cancer drug, while their sister cells are again not," says Bart Deplancke.
Testing Live-seq, the researchers showed that it can accurately identify ("stratify") diverse cell types and states without introducing major disturbances. As a proof-of-concept, they used their new platform to directly map the "trajectory" of individual immune cells (macrophages) before and after they became active, as well as adipose stromal cells—a type of stem cells—before and after they differentiated into fat cells. Finally, the team used Live-seq as a "transcriptomic recorder," which allowed them to predict how strongly—or weakly—an immune cell would react to an immunological challenge.
The work is now published in Nature. "Live-seq can address a broad range of biological questions by transforming scRNA-seq from an end-point to a temporal and spatial analysis approach," says Julia Vorholt.
More information: Bart Deplancke, Live-seq enables temporal transcriptomic recording of single cells, Nature (2022). DOI: 10.1038/s41586-022-05046-9. www.nature.com/articles/s41586-022-05046-9
Journal information: Nature
Provided by Ecole Polytechnique Federale de Lausanne