This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Scientists develop strategy to engineer artificial allosteric sites in protein complexes
According to a recently published research paper, an approach has been developed to create artificial allosteric sites (where by binding an effector molecule, activity at the distal active site is regulated) in protein complexes. This breakthrough research holds significant promise for a wide range of applications in industrial, biological, medical, and agricultural fields. The work was published in Nature Chemistry.
Protein complexes, such as hemoglobin and molecular motors, exert concerted functions through cooperative work between the subunits (constituent proteins in the protein complex). This orchestration is enabled by the allosteric mechanism.
Allosteric effect, regulation of function at an active site in a subunit by the binding of an effector molecule to an allosteric site in another subunit, was originally proposed in the 1960s and since then it has remained one of the most important topics in the biochemistry field.
A research team has developed a strategy for designing artificial allosteric sites into protein complexes to regulate a concerted function of a protein complex. "The creation of artificial allosteric sites into protein complexes has the potential to reveal fundamental principles for allostery and serve as tools for synthetic biology," said Nobuyasu Koga, a professor at the Osaka University.
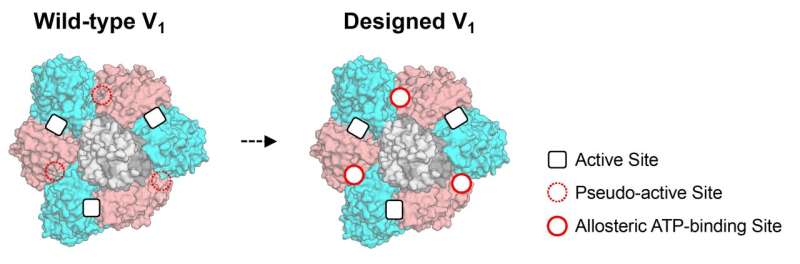
The research team hypothesized that allosteric sites in protein complexes can be created by restoring lost functions of the pseudo-active sites which are predicted to have been lost during evolution. Various protein complexes include subunits that have pseudo-active sites.
It has been reported that pseudo-active sites have an allosteric connection with active sites in other subunits. For example, a pseudo-active site in a subunit, which has lost ATPase activity but still exhibits ATP-binding ability, activates another subunit's active site upon binding to ATP. At the cellular level, ATP is the source of energy. ATPase describes the enzyme's ability to decompose ATP. Such studies support the idea that distinct allosteric sites can be created into protein complexes by engineering pseudo-active sites.
The research team used computational design to restore the lost ATP-binding ability of the pseudo-active site in the B-subunit of a rotary molecular motor, V1-ATPase. First, binding ability of the designed site was experimentally revealed by X-ray crystallography. "The X-ray structure indicated the binding site is successfully designed and integrated into the natural protein to have a function. I was amazed at the utility with high performance of protein design technology," said Mikio Tanabe, an associate professor at the Institute of Materials Structure Science.
Next, their single-molecule experiments with X-ray crystallography analyses revealed that the binding of ATP to the designed allosteric site boosts this V1's activity compared to the wildtype, and the rotation rate can be tuned by modulating the ATP's binding affinity. The team created cooperativity in the rotary motor.
Moreover, the team's designed allostery accelerated, not decelerated, the rotation, compared to the wildtype. "To our best knowledge, acceleration of a rotary molecular motor by protein engineering is a first time achievement. This is an exciting result in the field," said Ryota Iino a professor at the National Institutes of Natural Sciences.
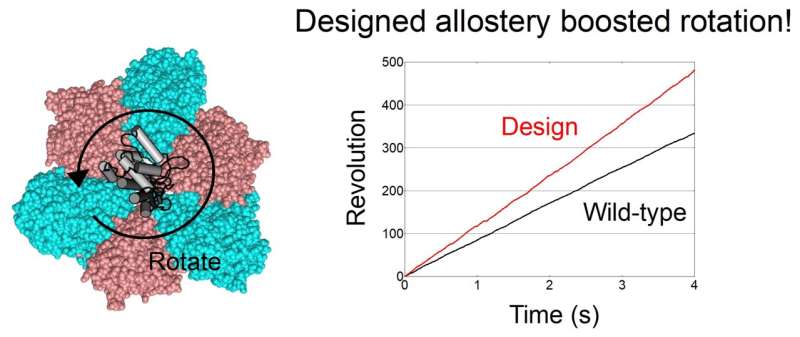
Pseudo-active sites are widespread in nature, and their approach shows promise as a means of programming allosteric control over concerted functions of protein complexes. In addition, the protein design method would enable not only to restore the lost function but also to design binding sites for other ligands.
"Our strategy enables us to create allosteric sites into various kinds of protein complexes in principle. Our next step is to create allosteric control for a variety of protein complexes by our strategy. Moreover, we will try to design novel functions into pseudo-active sites. One of our goals is to artificially control concerted functions of any protein complexes and to uncover the general mechanism of allostery. We hope allosteric control over concerted functions of protein complexes will open up new avenues in industrial applications of enzymes or biological, medical and agricultural fields," said Takahiro Kosugi an assistant professor at the National Institutes of Natural Sciences.
More information: Design of allosteric sites into rotary motor V1-ATPase by restoring lost function of pseudo-active sites, Nature Chemistry (2023). DOI: 10.1038/s41557-023-01256-4
Journal information: Nature Chemistry
Provided by National Institutes of Natural Sciences