This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Scientists discover new clues to cause of devastating coral disease
Biologists at The University of Texas at Arlington have published new findings from a study to learn how different coral species respond to a devastating disease and which species are more vulnerable.
The project examines the effects of stony coral tissue loss disease (SCTLD), which first appeared in 2014 in the waters around Florida and began spreading to the rest of the Caribbean in 2018-19. The research was conducted in the lab of Laura Mydlarz, UTA professor of biology, along with contributions from colleagues in the U.S. Virgin Islands and elsewhere.
The paper, "Stony coral tissue loss disease induces transcriptional signatures of in situ degradation of dysfunctional Symbiodiniaceae", is published in the May 22 edition of Nature Communications. Kelsey Beavers, a Ph.D. student in Mydlarz's lab, is lead author of the study.
SCTLD is a devastating, rapidly spreading disease characterized by rapid tissue loss and high mortality rates in coral. It has affected corals along the entire 350-plus miles of Florida's Coral Reef and in 22 Caribbean countries and territories, including the Virgin Islands and Puerto Rico. Corals are under stress and more susceptible to disease due to the effects of climate change.
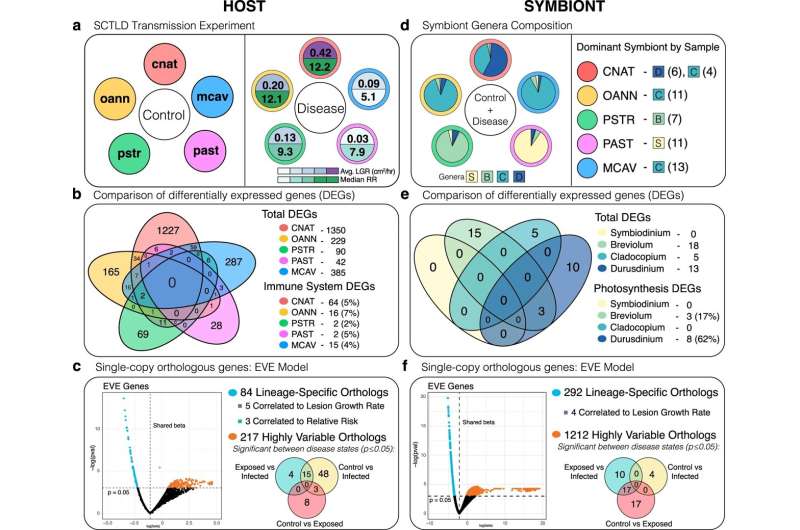
The team examined gene expression in five coral species in order to understand which species are more susceptible to the disease, which are affected more severely, and how fast the disease spreads in specific coral.
"We really wanted to figure out what are some conserved responses to the disease and whether that could give us an idea of what the disease is, because even after all this time since 2014, the field doesn't know what is causing stony coral tissue loss disease," Beavers said.
"We did see a difference in species susceptibility that provided this amazing framework for gene expression, and we found this new potential mechanism of this disease that is the coral getting rid of its own symbionts," Mydlarz said. "The symbionts provide the food and the energy for the coral."
A symbiont is an organism living in a mutually beneficial relationship with another organism. In this case, the symbiont is Symbiodiniaceae, which are algae that live inside coral. The coral provides a safe home for the Symbiodiniaceae, and the Symbiodiniaceae use sunlight to produce food for the coral.
The researchers found that SCTLD infection causes an increased expression of the gene rab7, which has been found in corals to be involved in a process called symbiophagy, where coral will digest dead or dysfunctional symbionts.
"Basically, this is the first disease where we've found that the coral may actually be mounting a response against this symbiont instead of just a general immune response," Beavers said. "This indicates that the symbionts that typically live happily inside the coral and give it energy could actually be the source of the disease."
Data gathered during the study showed that in addition to trying to digest their own symbionts to get rid of them, the corals are also undergoing some measure of starvation.
"We found lots of lines of evidence implicating the symbiont as the source, because the symbionts are also experiencing stress in their photosynthesis, either going haywire or just turning off," Beavers said. "Also, most likely as a consequence of that, we see genes indicative of starvation in the coral. So, if they're not getting food from their symbiont, it would make sense that they're having starvation responses."
One thing that stood out to the team was that they observed a high number of genes involved in antiviral immunity in the corals, and there has been some evidence in the field that one or more viruses could be involved in the disease.
"Our results show activation of antiviral immunity in the coral, which indicates that they're fighting some kind of viral infection as well," Beavers said. "We think with the symbiont involvement and the antiviral immune genes that it could be a virus of the symbiont. That's our current hypothesis, but we need to do more research to follow up on that."
Mydlarz noted that the severely high mortality rates among corals have prompted scientists to take drastic measures. This includes removing healthy corals from reefs before disease can strike them, and transporting them to aquariums, zoos or other facilities where they'll be safe. Some corals from Florida have even ended up at the Fort Worth Zoo.
"The positive aspect of this is we're closer to figuring out what's going on with this disease, which would mean we could predict it better for management purposes, like treating corals on the reef or moving corals," Mydlarz said.
"That's why we're doing this, because there's a huge field of people who are very passionate about coral, and everyone knows what's going to happen if we don't intervene," Beavers said.
Co-authors of the study include current and former members of Mydlarz's lab Emily Van Buren, Madison Emery, Nicholas MacKnight, and Bradford Dimos, as well as Marilyn Brandt from the University of the Virgin Islands, St. Thomas, and members of UVI's Center for Marine and Environmental Studies. Researchers from Mote Marine Laboratory in Florida, Rice University and Louisiana State University also contributed to the project.
More information: Kelsey M. Beavers et al, Stony coral tissue loss disease induces transcriptional signatures of in situ degradation of dysfunctional Symbiodiniaceae, Nature Communications (2023). DOI: 10.1038/s41467-023-38612-4
Journal information: Nature Communications
Provided by National Science Foundation