This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Uncovering hidden mitochondrial mutations in single cells
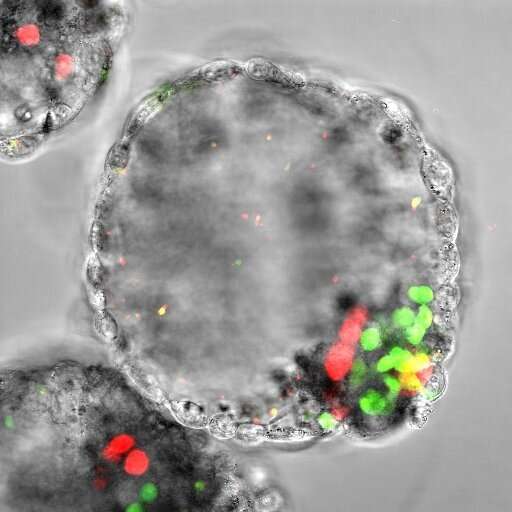
A high-throughput single-cell single-mitochondrial genome sequencing technology known as iMiGseq has provided new insights into mutations of mitochondrial DNA (mtDNA) and offers a platform for assessing mtDNA editing strategies and genetic diagnosis of embryos prior to their implantation.
An international team of researchers, led by KAUST stem cell biologist Mo Li, has now quantitatively depicted the genetic maps of mtDNA in single human oocytes (immature eggs) and blastoids (stem cell-based synthetic embryos). This has revealed molecular features of rare mtDNA mutations that cause maternally inherited diseases.
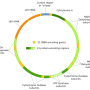
Mitochondria play a crucial role in cellular communication and metabolism. Human mtDNA is a circular genome containing 37 genes, encoding 13 proteins and a noncoding D-loop region. Heteroplasmic mutations, inherited from egg cells, can cause congenital diseases, like maternally inherited Leigh syndrome, and are associated with late-onset complex diseases.
"Next-generation sequencing has been used to sequence mtDNA and implicated heteroplasmic mutations as significant contributors to metabolic disease. Yet the understanding of mtDNA mutations remains limited due to the constraints of traditional sequencing technologies," says lead author Chongwei Bi.
"Our new iMiGseq method is significant because it enables complete sequencing of individual mtDNA in single cells, allowing for unbiased, high-throughput base-resolution analysis of full-length mtDNA," says Bi. iMiGseq resolves several key questions in the field.
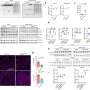
Using third-generation nanopore sequencing technology, the researchers have characterized mtDNA heteroplasmy in single cells and described the genetic features of mtDNA in single oocytes. They have examined mtDNA in induced pluripotent stem cells derived from patients with Leigh syndrome or neuropathy, ataxia or retinitis pigmentosa (NARP). This has revealed complex patterns of pathogenic mtDNA mutations, including single nucleotide variants and large structural variants. "We were able to detect rare mutations with frequencies far below the traditional detection threshold of 1%," says Mo Li.
In another experiment using the new technology, iMiGseq revealed the potential risks of unexpected large increases in the frequency of off-target mutations, known as heteroplasmy, in a mitochondrial genome editing method called mitoTALEN—a genome editing tool that cuts a specific sequence in mitochondrial DNA. It is used to cut a mutation that causes mitochondrial encephalomyopathy and stroke-like episodes syndrome in patient-derived induced pluripotent stem cells.
Both studies are published in Nucleic Acids Research.
"This highlights the advantages of full-length mtDNA haplotype analysis for understanding mitochondrial DNA heteroplasmy change; other distant mtDNA genetic variants may be unintentionally affected by the editing of a genetically linked disease-relevant mutation and there is a need for ultrasensitive methods to assess the safety of editing strategies," says Li.
The researchers also used iMiGseq to analyze single human oocytes from healthy donors and single human blastoids, synthetic embryos made from stem cells, to identify rare mutations undetectable with conventional next-generation sequencing. These low-level heteroplasmic mutations, potentially inherited through the female germline, are linked to mitochondrial diseases and cancer.
The iMiGseq method provides a novel means to accurately depict the complete haplotypes of individual mtDNA in single cells, offering an ideal platform for explaining the cause of mitochondrial mutation-related diseases, evaluating the safety of various mtDNA editing strategies and unraveling the links between mtDNA mutations, aging and the development of complex diseases.
More information: Chongwei Bi et al, Quantitative haplotype-resolved analysis of mitochondrial DNA heteroplasmy in Human single oocytes, blastoids, and pluripotent stem cells, Nucleic Acids Research (2023). DOI: 10.1093/nar/gkad209
Chongwei Bi et al, Single-cell individual full-length mtDNA sequencing by iMiGseq uncovers unexpected heteroplasmy shifts in mtDNA editing, Nucleic Acids Research (2023). DOI: 10.1093/nar/gkad208
Journal information: Nucleic Acids Research