This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Sizing them up: An algorithm to accurately quantify rapeseed silique morphology
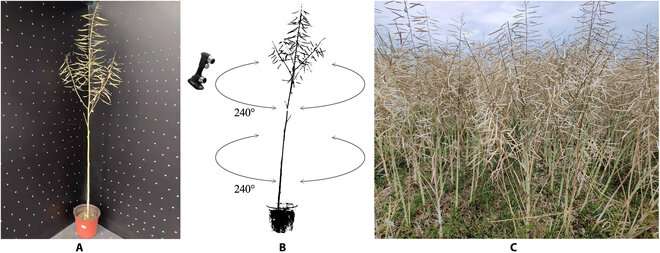
Rapeseed or oilseed rape (Brassica napus L.) is an important crop cultivated worldwide for its oil-rich seeds. The rapeseed silique is an organ that plays a role in photosynthesis, sends developmental signals to maturing seeds, and provides a capsule that harbors the seeds.
High-yield rapeseed varieties have both a high number and optimal morphology–the form and structure–of siliques. In this regard, rapeseed genotype and cultivation method directly influence the number of siliques that a plant produces. Thus, accurately quantifying silique development parameters is critical for predicting yield and identifying high-yield varieties.
Traditionally, silique developmental parameters–silique length (SL) and silique number (SN)–have been manually quantified, making the process inherently invasive, inaccurate, and time-consuming. Two-dimensional (2D) and three-dimensional (3D) agro-optics have been able to circumvent these difficulties but come with their limitations. 2D imaging methods are less robust and cannot gather complete spatial information.
On the contrary, 3D imaging parameter tuning is made tedious by the complexities of plant structure, and the clustering algorithms used for phenotyping–the process of recording the observable traits of an organism–often cannot differentiate between plant skeletons and plant organs. Now researchers in China have developed a new algorithm that uses 3D imaging data to deliver non-invasive phenotyping. The team's findings were recently published in Plant Phenomics.
"We wanted to develop a high-throughput method that better predicted yield. Our objective was to design a tool that delivered a skeleton model of oilseed rape, accurately segmented siliques, and gathered morphological data of siliques," explains Professor Haiyan Cen, the team lead and current Vice Dean in the College of Biosystems Engineering and Food Science, Zhejiang University.
The team's approach involved combining two types of algorithms–a skeletonization algorithm and a hierarchical segmentation algorithm–to accurately separate siliques from the whole plant using 3D imaging data collected by a laser scanner. By incorporating distance, angle, and direction information of the individual components of the skeleton, the 3D image data was used to create a detailed representation of the plant in a cartesian plane.
The skeletonization with hierarchical segmentation (SHS) algorithm was tested for its capacity to automatically quantify the silique volume (SV), SL, and SN from oilseed rape plants cultivated in greenhouses and the field.
"Our algorithm delivered a high degree of accuracy when extracting morphological data from rapeseed siliques. Its predictions for SN, total SL, and total SV showed very strong and statistically significant correlations with the actual yield of the plant," says Prof. Cen, when elaborating on the key abilities of the team's new algorithm.
The SHS performed well in its estimation of silique segmentation and phenotypic extraction too, with high correlations achieved for both SN and total SL. Another key finding was that the SHS could even differentiate between rapeseed plants based on their branching architecture into plants of few-branch broom shape (FBBS), plants of multibranch broom shape (MBBS), and plants of multibranch cylinder shape (MBCS). Here too, the SHS was able to detect statistically significant differences between the three types of branching patterns.
Prof. Cen and her team are excited about the future directions of their research. They believe that given its high degree of accuracy in yield estimation and phenotyping analyses, their non-invasive SHS has the potential to provide technical support for oilseed rape breeding operations the world over.
More information: Zhihong Ma et al, Phenotyping of Silique Morphology in Oilseed Rape Using Skeletonization with Hierarchical Segmentation, Plant Phenomics (2023). DOI: 10.34133/plantphenomics.0027
Provided by NanJing Agricultural University