This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers develop rapid Salmonella test to improve safety, reduce waste and lower costs
McMaster University researchers have developed a rapid and inexpensive test for Salmonella contamination in chicken and other food—one that's easier to use than a home COVID test.
The test, described in a new paper in the journal Angewandte Chemie International Edition, could improve food safety, reduce the cost of processing fresh poultry and other foods, and help to limit broad recalls to batches that have specifically been identified as contaminated.
The researchers have shown that the test provides accurate results in an hour or less without the need for accessories or a power source, compared to today's monitoring through lab cultures, which require at least a full day to produce results.
Once scaled up and made available commercially, the new test could be a significant boon to poultry processors, for whom Salmonella is among the most significant contamination risks. The test would also be beneficial for ensuring the safe processing of other foods that are particularly vulnerable to Salmonella, such as eggs, dairy products and ground beef.
A single major poultry processor performs tens of thousands of Salmonella lab tests each year. Reducing or even eliminating the need for overnight lab cultures would represent significant savings and make it easier to identify contamination earlier in the process.
"Anyone can use it right in the setting where food is being prepared, processed or sold," says co-author Yingfu Li, a professor of Biochemistry and Chemical Biology who leads McMaster's Functional Nucleic Acids Research Group. "There's a balance between cost, convenience and need. If it's cheap, reliable and easy, why not use it?"
Protecting the public from Salmonella is a high priority for food producers, retailers and regulators alike, since Salmonella is one of the most common and serious forms of food-borne infection, causing 155,000 deaths globally every year.
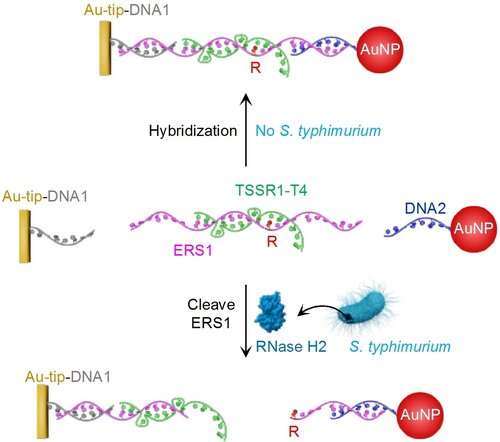
What makes the test work is a new synthetic nucleic acid molecule, developed at McMaster. For the test, the molecule is sandwiched between microscopic particles such as gold.
The test platform lines the inside of the tip of a pipette and begins to work when a liquefied sample of the food being tested is drawn inside the tube.
If Salmonella bacteria are present, they cut through the particles, allowing the molecule to escape.

When the solution is dropped onto a paper test strip, the presence of Salmonella shows as a visible shade of red, thanks to a new form of biosensor, also created by the McMaster team. The greater the concentration of Salmonella, the brighter the color appears.
"Using these tests is easier than using a COVID test, which so many people are already doing," says co-author Carlos Filipe, chair of McMaster's Department of Chemical Engineering. "For this to be as effective and useful as possible, it has to be easy to use."
The new technology has been developed with support from the non-profit research organization Mitacs, and Toyota Tsusho Canada Inc., an indirect subsidiary of Toyota Tsusho Corporation in Japan, which plans to develop the innovation for commercial use.
The research is part of an ongoing, broader effort to establish McMaster and its Global Nexus for Pandemics and Biological Threats as a center for the development of real-time sensors, pathogen-repellent materials and other products that improve food safety.
"This is very important to us in the development of our food-testing program," says co-author Tohid Didar, an associate professor of Mechanical Engineering and Canada Research Chair in Nano-biomaterials. "Being able to create a test that is both easy to use and which produces a readily visible color within an hour is significant."
Li, Didar and Filipe authored the paper with postdoctoral research fellow Jiuxing Li, Ph.D. student and Vanier Scholar Shadman Khan, and research associate Jimmy Gu.
Reducing illness and food waste aligns with Toyota Tsusho Canada's values, explained Toyota Tsusho Canada Inc. President Grant Town.
"Our goal is to help bring proven research from the lab to the marketplace, where it can benefit society," Town says. "Reducing the risk of illness while also cutting food waste will benefit everyone, and Toyota Tsusho Canada sees this as a great opportunity."
More information: Jiuxing Li et al, A Simple Colorimetric Au‐on‐Au Tip Sensor with a New Functional Nucleic Acid Probe for Food‐borne Pathogen Salmonella typhimurium, Angewandte Chemie International Edition (2023). DOI: 10.1002/anie.202300828
Journal information: Angewandte Chemie International Edition
Provided by McMaster University