This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Study compares de novo proteins with randomly produced proteins
Proteins are components of every cell. How they have changed in the course of evolution for the purpose of taking on new functions in the body, has long been a subject of research. The fact that proteins can emerge practically out of nothing—out of a new DNA structure emerging at random, in previously non-coding parts of the genome—has only been established relatively recently and has not been the object of much investigation in comparison with the "traditional" evolutionary processes.
A team of Czech and German researchers headed by biochemist Dr. Klára Hlouchová from the University of Prague and bioinformatics specialist Prof. Dr. Erich Bornberg-Bauer from the University of Münster have now, for the first time, carried out experiments comparing de novo proteins with computer-generated proteins as regards their stability and solubility—and have been able to demonstrate small but significant differences between them. The study has been published in the current issue of Nature Ecology and Evolution.
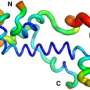
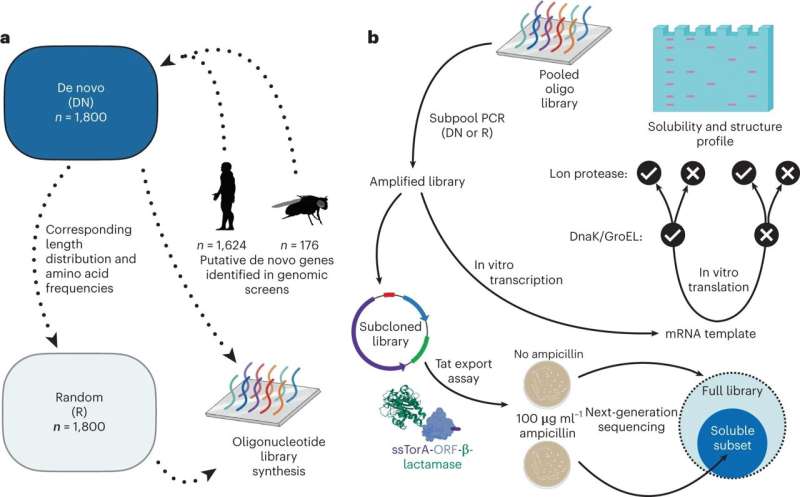
The team compared two sorts of proteins: 1,800 candidates for de novo proteins occurring in fruit flies and humans, where they are located in non-genic parts of the genome in the form of DNA, and randomly computer-generated proteins. While predictions relating to structure—which the researchers carried out with the aid of various computer programs—made very similar classifications of both classes of proteins, the investigations carried out in the lab showed small differences which the predictions did not show up.
For example, the de novo proteins in the lab experiments displayed on average a slightly higher solubility, based on the so-called secondary structure. "What we also found is that, in spite of their origins being so young, de novo proteins can be better integrated into the cell than we would have expected from proteins emerging at random," says lead author Brennen Heames from Münster. "These results point to natural selection occurring in the early phase of these proteins' emergence."
Margaux Aubel, co-author from the Münster group, adds, "Our results are particularly useful for basic research in the field of de novo evolution. But in our dataset there are many human de novo proteins which we investigated for solubility and for their ability to aggregate. This latter ability plays a role in a variety of diseases, and some studies have already shown that de novo proteins may also be associated with diseases. Perhaps our results will help us to learn more about the role of these hitherto under-researched proteins in the development of diseases."
Previous studies have often looked at de novo evolution from a purely theoretical angle, examining large datasets. Experimental studies, on the other hand, mostly investigate individual de novo proteins. Although a comparison of de novo proteins with randomly generated sequences has already been undertaken in theoretical studies, it has never been verified in experiments. Because the de novo proteins are relatively young, occurring in non-coded DNA exposed to little or no evolutionary pressures, these proteins are more readily comparable to randomly generated proteins than to old established proteins.
The team used computer programs to predict proteins' properties. The researchers produced the proteins required for experimental analysis, scrutinizing them by means of mass spectrometry. In a further round of experiments, they added a protein-degrading enzyme which enabled them to test how many of the proteins were actually degraded and then to draw conclusions regarding their stability. In order to investigate the proteins' solubility, the team used a molecular transport mechanism of the Escherichia coli bacterium as an indicator. The soluble proteins thus identified were more closely defined through next generation DNA sequencing.
More information: Brennen Heames et al, Experimental characterization of de novo proteins and their unevolved random-sequence counterparts, Nature Ecology & Evolution (2023). DOI: 10.1038/s41559-023-02010-2
Journal information: Nature Ecology & Evolution
Provided by University of Münster