March 9, 2023 report
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Using peptides instead of DNA to tag molecules to speed up drug discovery
A team of chemists at MIT has found that using peptides instead of DNA fragments to tag small molecules can speed up the drug discovery process. Their research is published in the journal Science.
Developing drugs to treat or cure human ailments is generally a long and expensive process. Quite often, it starts with chemists exposing proteins with known properties to large numbers of small molecules, hoping to find one that binds tightly.
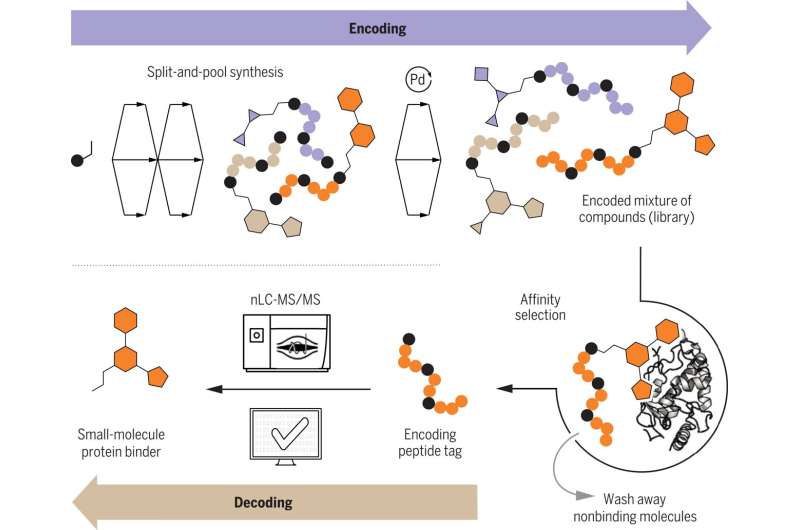
Part of this process involves tagging the small molecules beforehand to identify them after binding takes place. Typically, this has involved using short fragments of DNA. The downside to using DNA, however, is its sensitivity and incompatibility with some chemicals and catalysts. In this new effort, the researchers sought to find a replacement tag to make the work of finding new drugs more efficient.
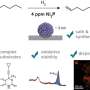
In their search, the researchers tested using peptides because they are known to be useful under a broader range of conditions. They also found that they were able to speed up the decoding process by using mass spectrometry.
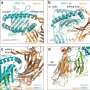
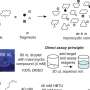
To use peptides as tags, the chemists first immobilized the small molecule under study using a resin. This created a link between the small molecule and an amino acid. Reactions were then performed iteratively. To create a library, the resin was cut into smaller portions, with each treated differently. After the reactions were complete, the resin was recombined and split again to allow for the next run.
This approach allowed for testing large numbers of small molecules at a much faster pace than is currently possible using DNA as tags. After identifying likely candidates via mass spectrometer, the researchers used software to convert the spectra into a unique peptide sequence. The team also wrote a Python program to process the data.
The researchers note that in addition to speeding up the process of finding new drugs, replacing DNA tags with peptide tags allows for creating longer tag strings—DNA only allows for four letters. Peptides, on the other hand, allow for 16.
More information: Simon L. Rössler et al, Abiotic peptides as carriers of information for the encoding of small-molecule library synthesis, Science (2023). DOI: 10.1126/science.adf1354
Journal information: Science
© 2023 Science X Network