This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Computer model IDs roles of individual genes in early embryonic development
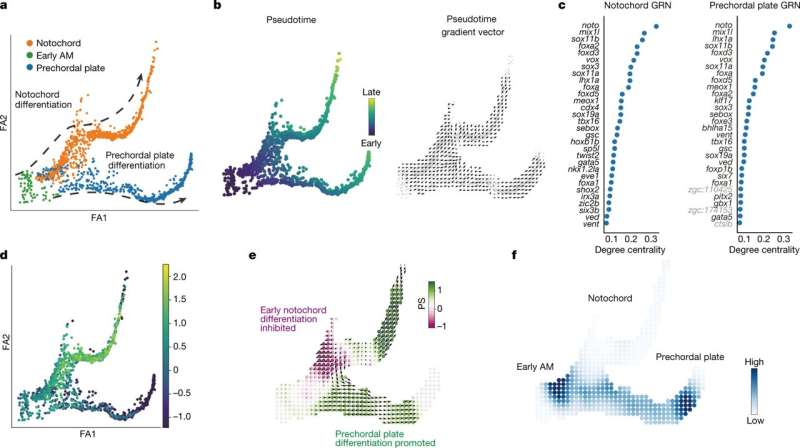
Computer software developed at Washington University School of Medicine in St. Louis can predict what happens to complex gene networks when individual genes are missing or dialed up more than usual. Such genetic networks play key roles in early embryonic development, guiding stem cells to form specific cell types that then build tissues and organs. Mapping the roles of single genes in these networks is key to understanding healthy development and finding ways to regrow damaged cells and tissues. Likewise, understanding genetic errors could provide insight into birth defects, miscarriage or even cancer.
Such genetic experiments—typically conducted in the laboratory in animal models such as mice and zebrafish—have been a mainstay of developmental biology research for decades. Much can be learned about a gene's function in animal studies in which a gene is missing or overexpressed, but these experiments are also expensive and time-consuming.
In contrast, the newly developed software called CellOracle—described Feb. 8 in the journal Nature—can model hundreds of genetic experiments in a matter of minutes, helping scientists identify key genes that play important roles in development but that may have been missed by older, slower techniques. CellOracle is open source, with the code and information about the software available here.
"The scientific community has collected enough data from animal experiments that we now can do more than observe biology happening—we can build computer models of how genes interact with each other and predict what will happen when one gene is missing," said senior author Samantha A. Morris, Ph.D., an associate professor of developmental biology and of genetics. "And we can do this without any experimental intervention. Once we identify an important gene, we still need to do the lab experiments to verify the finding. But this computational method helps scientists narrow down which genes are most important."
CellOracle, which was included in a recent technology feature in the journal Nature, is one of a number of relatively new software systems designed to model insights into cellular gene regulation. Rather than simply identify the networks, CellOracle is unique in its ability to let researchers test out what happens when a network is disrupted in a specific way.
Morris and her team harnessed the well-known developmental processes of blood cell formation in mice and humans and embryonic development in zebrafish to validate that CellOracle works properly. Their studies, in collaboration with the lab of co-author and zebrafish development expert Lilianna Solnica-Krezel, Ph.D., the Alan A. and Edith L. Wolff Distinguished Professor and head of the Department of Developmental Biology, also uncovered new roles for certain genes in zebrafish development that had not previously been identified.
And in a related paper published in the journal Stem Cell Reports, Morris and her colleagues used CellOracle to predict what happens when certain genes are dialed up beyond their usual expression levels.
"We found that if we dialed up two specific genes, we can transform skin cells into a type of cell that can repair damaged intestine and liver," Morris said. "In terms of regenerative medicine, these predictive tools are valuable in modeling how we can reprogram cells into becoming the types of cells that can promote healing after injury or disease."
According to Morris, most laboratory methods for converting stem cells into different cell types, such as blood cells or liver cells, are inefficient. Maybe 2% of the cells arrive at the desired destination. Tools like CellOracle can help scientist identify what factors should be added to the cocktail to guide more cells into the desired cell type, such as those capable repairing the gut and liver.
At present, CellOracle can model cell identity in more than 10 different species, including humans, mice, zebrafish, yeast, chickens, Guinea pigs, rats, fruit flies, roundworms, the Arabidopsis plant and two species of frog.
"We get a lot of requests to add different species," Morris said. "We're working on adding axolotl, which is a type of salamander. They are cool animals for studying regeneration because of their ability to regrow entire limbs and other complex organs and tissues."
More information: Kenji Kamimoto et al, Dissecting cell identity via network inference and in silico gene perturbation, Nature (2023). DOI: 10.1038/s41586-022-05688-9
Kenji Kamimoto et al, Gene regulatory network reconfiguration in direct lineage reprogramming, Stem Cell Reports (2022). DOI: 10.1016/j.stemcr.2022.11.010
Journal information: Stem Cell Reports , Nature