This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
'Snapshots' of translation could help us investigate cellular proteins
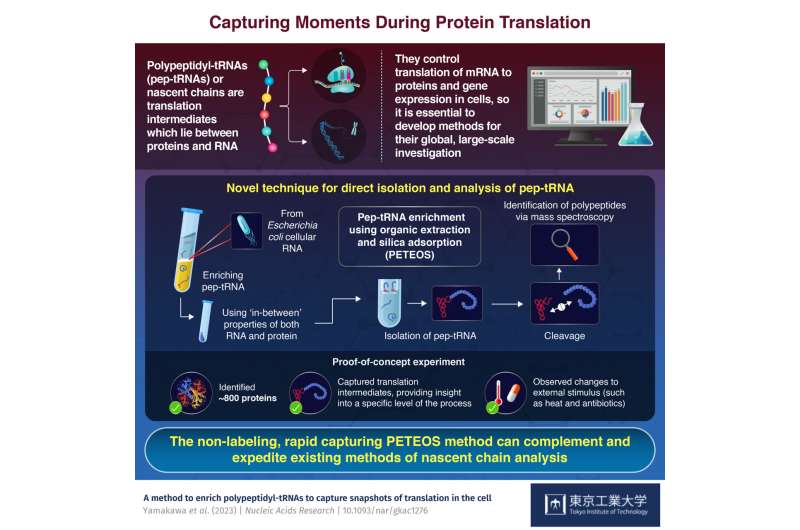
Advances in molecular biology have revealed that pep-tRNAs—nascent polypeptides inside the ribosome that are covalently attached to transfer RNA—are involved in myriad cell functions, including gene expression. All proteins exist as pep-tRNAs at some point, and studying these translation intermediates is vital as they possess properties of both RNA and protein, and can help researchers better understand the specifics of translation.
Depending on stimuli and/or stresses, translational regulation is very rapid and spans initiation, elongation, and elongation pausing. Garnering deeper insights into the process of translation therefore requires a suitable method to process pep-tRNAs in large quantities. These nuances have fueled the development of molecular tools to investigate cellular translation.
Presently, the two primary approaches being used to survey cellular translation leverage vastly different strategies. The first, ribosome profiling, a powerful deep-sequencing technology, monitors translation by targeting ribosome-protected mRNA fragments produced from RNase digestion. Unfortunately, this approach is inherently unable to capture pep-tRNAs, and is time-consuming, expensive, and data intensive.
The alternative relies on proteomics with liquid chromatography-tandem mass spectroscopy (LC-MS/MS) and requires using unusual amino acids to label polypeptides. However, such proteomics approaches are time-intensive, and, most importantly, do not capture the translation elongation status of the cell, as pep-tRNAs aren't directly targeted.
To navigate around these limitations in "pep-tRNA-ome" technology, a team of scientists from Tokyo Institute of Technology (Tokyo Tech), led by Prof. Hideki Taguchi, has developed PETEOS. Their findings have now been published in Nucleic Acids Research.
Regarding the main advantages of their work, Prof. Taguchi explains, "This non-labeling methodology is unique in that it specifically enriches and rapidly captures pep-tRNAs while still complementing conventional translatome analyses platforms."
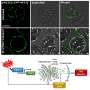
PETEOS has four steps. First, RNA is enriched using organic solvent extraction and then isolated on a silica column. Next, the target polypeptides in the isolated pep-tRNAs are separated from their ribonucleotides using a combination of high pH and temperature. Then, the liberated polypeptides are subjected to enzymatic digestion and cleaved. And lastly, these polypeptides are identified using shotgun LC-MS/MS proteomics.
"As a proof-of-concept, we were able to identify nearly 800 E. coli proteins that were derived from pep-tRNA using PETEOS. Importantly, these proteins were N-termini-enriched, which underscores that the method really does capture the intermediate pep-tRNA pool during translation," says Prof. Taguchi, speaking of the significance of their new method.
The team was also able to show that PETEOS could capture "screenshots" of the nascentome—the pool of nascent pep-tRNAs present at any given time—when E. coli cells were subjected to heat shock and antibiotic treatments. PETEOS outperformed conventional methods in terms of the capacity to capture acute protein expression under heat stress, and the rearrangement of the nascentome following exposure to different antibiotics.
PETEOS does have a few limitations, including a difficulty in analyzing low abundance pep-tRNAs using LC-MS/MS, limited quantitative capacity because of the acidic phenol/chloroform step, no codon-level resolution, and difficulty capturing the C-terminal end of the growing polypeptide. However, some of these issues exist for conventional approaches as well and can be overcome by improving the C-terminal peptide enrichment efficiency of this process.
The team believes there is tremendous potential to optimize their protocol moving forward. Prof. Taguchi concludes, "Despite the limitations, we're confident PETEOS has potential as it enables analyses that current translatome methods simply cannot. Most importantly, it can be applied to any organism." With more research in this field, we will deepen our understanding of protein synthesis.
More information: Ayako Yamakawa et al, A method to enrich polypeptidyl-tRNAs to capture snapshots of translation in the cell, Nucleic Acids Research (2023). DOI: 10.1093/nar/gkac1276
Journal information: Nucleic Acids Research
Provided by Tokyo Institute of Technology