Scientists decrypt the 'mechanical code' of DNA
A new study has deciphered the mechanical code of DNA to reveal previously unknown ways in which nature encodes biological information in DNA sequence.
Led by Durham University, UK, an international team of researchers used a next-generation DNA-sequencing based technology called loop-seq, which they had developed, to show that the local sequence of bases along a region of DNA determines the local bendability of DNA.
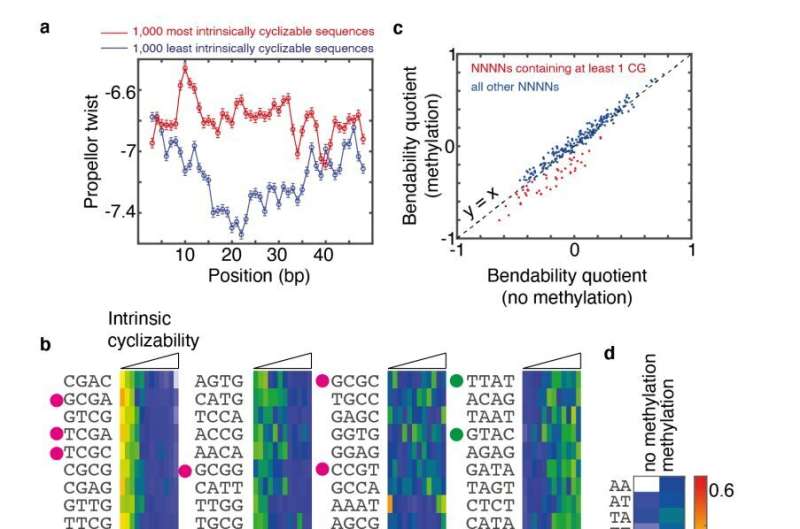
Via a large number of measurements, coupled with computational analysis and machine learning, they determined the mechanical code, i.e., the mapping between local sequence and the local deformability of DNA.
Additionally, the researchers found that the mechanical code of DNA can be modified by "methylation," which is a known chemical modification that DNA bases are routinely subject to at various stages in an organism's development. Aberrant methylation has been linked to several cancers.
The discovery that methylation alters the mechanical code presents the possibility that biological development programs, or diseases such as cancer, could be achieving a part of their effects on cells by altering the information encoded via the mechanical code.
The research was carried out along with colleagues from Johns Hopkins University, U.S., Barcelona Institute of Science and Technology, Spain, and the University of Barcelona, Spain. It has been published in the journal Nature Structural & Molecular Biology.
Lead author of the study, Dr. Aakash Basu of Durham University, said, "DNA is a book containing instructions that cells need to survive. But it's a very special kind of book, where your ability to turn a page, repair a tear in the page, or fold a page, depend on the words written on the page. This is because in the book of DNA, those words somehow also control the mechanical properties of the paper."
They point out that it is well known that, reading, copying, packaging, and repairing the genetic information stored in the sequence of bases (the As, Ts, Gs, and Cs) along DNA routinely involves processes that require local mechanical deformations of DNA.
The researchers provide evidence that in diverse organisms ranging from mammals to bacteria, nature and evolution has taken advantage of the mechanical code to locally control DNA deformability, and thus in turn, control critical biological processes that require mechanical distortions of DNA.
The researchers expect this knowledge to guide future therapeutic and bioengineering developments.
More information: Aakash Basu, Deciphering the mechanical code of the genome and epigenome, Nature Structural & Molecular Biology (2022). DOI: 10.1038/s41594-022-00877-6. www.nature.com/articles/s41594-022-00877-6
Journal information: Nature Structural & Molecular Biology
Provided by Durham University