CRISPR insight: How to fine-tune Cas protein's grip on DNA
At the heart of every CRISPR reaction, whether naturally occurring in bacteria or harnessed by CRIPSR-Cas gene editing technology, is a strong molecular bond of a Cas protein via a guide RNA to its target site on DNA. It's like a nanoscale ski binding.
"There's a balance between stably bound and coming off at the right time," said Michelle Wang, the James Gilbert White Distinguished Professor of the Physical Sciences and Howard Hughes Medical Institute Investigator in the College of Arts and Sciences. "What we really want is the ability to modulate the affinity. That gives us the possibility of fine-tuning the gene editing potential."
A Cas protein binding can't be too transient, according to Porter Hall, a biophysics doctoral candidate in the Wang Lab and the lead author of the publication. If it can't stably bind the target region of the DNA, precise gene editing may not be efficient, potentially leading to off-target effects. "But if the protein stays there forever, then the gene editing process cannot be completed," Hall said.
Examining the precise, molecular-level mechanisms involved in Cas binding to DNA, Wang and colleagues give the first mechanistic explanation of how a motor protein (RNA polymerase) removes a bound dCas, a version of Cas engineered to recognize a DNA sequence without performing a cut.
This insight reveals how to tune Cas removal, contributing to future CRISPR applications.
"Polarity of the CRISPR Roadblock to Transcription" published Dec. 5 in Nature Structural & Molecular Biology. Other contributors are lab members James Inman, Robert Fulbright and Tung Le, along with collaborators Guillaume Lambert, assistant professor in applied and engineering physics, Cornell Engineering, and Joshua Brewer and Seth Darst from the Rockefeller University.
"To fully realize the potential of CRISPR technology, it is crucial to obtain an in-depth mechanistic understanding of Cas binding stability," the researchers wrote. "This work highlights the importance of the R-loop in dCas binding stability and provides valuable mechanistic insights for broad applications of CRISPR technology."
The Wang Lab investigates how motor proteins move as they travel along DNA strands, carrying out vital biological processes.
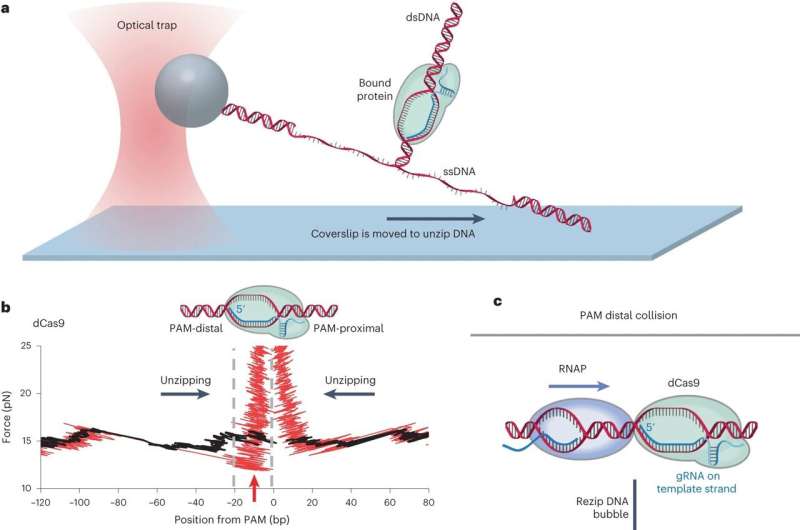
The motor protein RNA polymerase exerts force on "roadblocks" as it carries out its function of gene expression, copying DNA to RNA, Wang said. In this study, the roadblock was endonuclease-deficient Cas (dCas).
Previously, using nanophotonic tweezers, the researchers mechanically separated the two DNA strands to map out where the bound dCas protein is on the DNA. They call this the DNA unzipping mapper.
Previous research established that removal of dCas by a motor protein is possible only from one side (a polarity). Using the unzipping mapper for the current study, the Cornell researchers discovered why: because RNA polymerase can collapse the loop formed between the guide RNA and the target DNA (called the "R-loop") of a bound dCas only from one side, the side distal (or distant) from the PAM (protospacer adjacent motif), a short DNA sequence 2-6 base pairs long, that follows the DNA region targeted for cleavage.
Once the researchers describe how the mechanism works, they also show how to tune the dCas R-loop stability by modifying the guide RNA.
"We hope that fundamental knowledge of how Cas proteins work can ultimately lead to more efficient gene editing and broader applications of the CRISPR technology," Wang said.
More information: Porter M. Hall et al, Polarity of the CRISPR roadblock to transcription, Nature Structural & Molecular Biology (2022). DOI: 10.1038/s41594-022-00864-x
Journal information: Nature Structural & Molecular Biology
Provided by Cornell University