New insights into effects of membrane proteins on plant growth
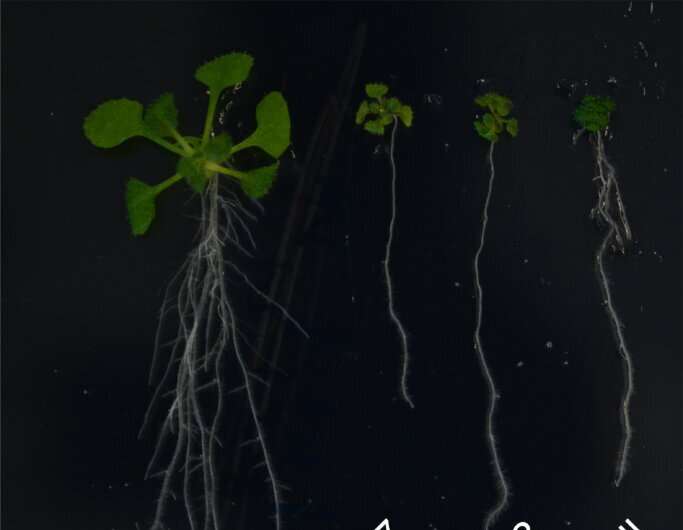
Scientists have revealed two enzymes that regulate protein degradation of proteins in the cell membrane of plants, and established the roles they play in plant growth and development.
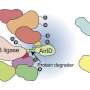
Ubiquitin is a small regulatory protein found in most cells of eukaryotic organisms. It is vital to altering the function of other proteins, and is most notably involved in protein degradation and protein localization within the cell. Ubiquitin regulates these functions by attaching to and detaching from the target protein, a process called ubiquitination.
Scientists from Japan, Belgium and the U.S., led by Associate Professor Takeo Sato at Hokkaido University, have discovered the first examples of deubiquitinating enzymes in plants that act on proteins in the cell membrane of the cell. Their findings, published in the journal EMBO reports, detail the function of these proteins.
The addition of ubiquitin to target proteins is catalyzed by enzymes called ubiquitin ligases, and the reverse process, removal of ubiquitin from target proteins, is catalyzed by deubiquitinating enzymes (DUBs). It was not known which plant DUBs were able to directly remove ubiquitin from membrane proteins. This lack of knowledge meant that the regulation of membrane protein stability was not fully understood.
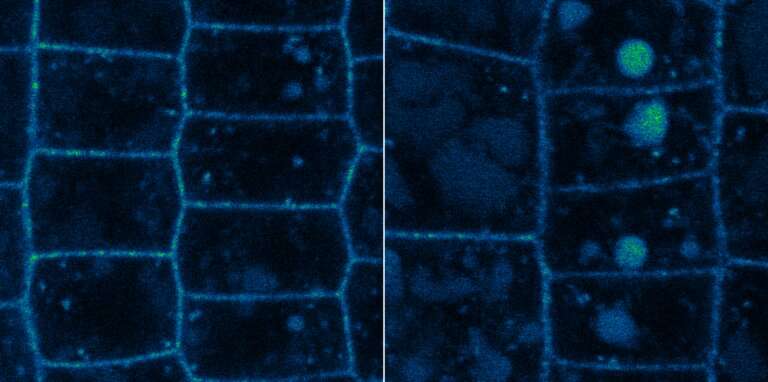
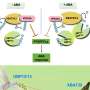
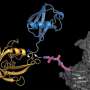
The team demonstrated that two of the Arabidopsis thaliana DUBs, UBP12 and UBP13, directly target the cell membrane-localized plant hormone receptor known as BRI1.
BRI1 is vital for the detection of brassinosteroids (BRs), steroidal phytohormones that are essential for growth and development. Typically, when BRI1 detects BR, it triggers a pathway that regulates gene expression in the nucleus. The abundance of cellular BRI1 is crucial to precisely mediate BR signal, although the regulatory mechanism for how BRI1 abundance is optimized (fine-tuned) in the cells was unclear. The scientists demonstrated that UBP12 and UBP13 de-ubiquitinated and stabilized BRI1.
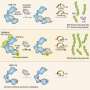
In their experiments, they showed that Arabidopsis thaliana plants that lacked the ability to express UBP12 and UBP13 were severely dwarfed and distinctly less sensitive to BRs. When a mutant BRI1—which could not be ubiquitinated—was introduced into these UBP12 and UBP13-deficient plants, growth defects were partially restored. Specifically, UBP12 and UBP13 target and act on ubiquitinated BRI1.
The study showed that UBP12 and UBP13 are key regulators of plant growth via their interaction with BRI1. The study also provided insight into how membrane protein stability is maintained; however, further research is required to fully understand the dynamics of BRI1 within the cell. Finally, the findings provide evidence that plant DUBs have roles similar to those of mammalian DUBs.
More information: Yongming Luo et al, Deubiquitinating enzymes UBP12 and UBP13 stabilize the brassinosteroid receptor BRI1, EMBO reports (2022). DOI: 10.15252/embr.202153354
Journal information: EMBO Reports
Provided by Hokkaido University