Human membrane proteins strike evolutionary balance
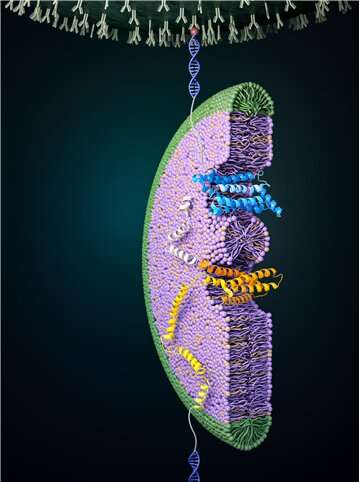
Cells are compartmentalized by membranes, and proteins present in these membranes play an important role in transporting cellular information. For proper function of these proteins to occur, a tertiary protein structure must be formed through the correct folding process. In a study published in Nature Chemical Biology, the folding process of a glucose transporter—a complex membrane protein—was identified for the first time using single-molecule magnetic tweezers.
Using the endoplasmic reticulum (ER) membrane protein complex (EMC) and a lipid molecule with a specific structure, the folding pathway of a glucose transporter was fully elucidated in a physiological environment. Through bioinformatics, it was also discovered that the membrane protein's structure-forming ability and its capacity to transport glucose had to have struck a balance throughout its evolutionary history.
Although the structures of many membrane proteins—including glucose transporters—have already been revealed through recent advances in structural biology such as cryo-electron microscopy, the folding pathway in which the structures of these membrane proteins is formed remains almost completely unknown. Back in 2019, the research team reported in Science that the folding pathways of membrane proteins can be revealed using magnetic tweezers, marking the first time in the world that the folding pathway of a membrane protein was revealed.
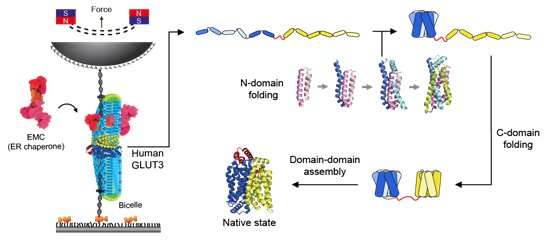
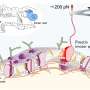
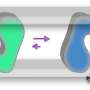
Magnetic tweezers can be used to apply force to a single protein, thereby completely unfolding a protein's structure. In addition, if the applied force is then lowered, it is possible to observe the process of the released protein refolding back into its original folded shape (Figure 1,2).
Glucose transporter proteins, as the name suggests, are a group of proteins that possess a pathway through which glucose can pass. Glucose transport pathways are functionally essential, but these pathways also act as obstacles in forming the tertiary structures of membrane proteins. Cells have various helpers to solve these difficulties. In this study, it was found that the EMC—a type of protein chaperone—as well as uniquely-structured lipid molecules work in conjunction in helping the glucose transporter in its structure formation.
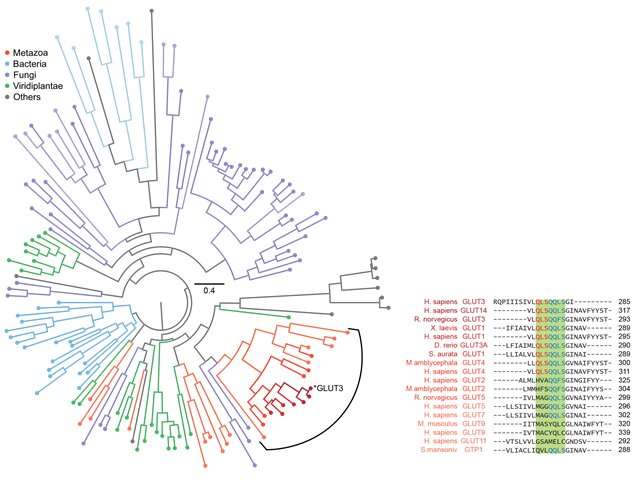
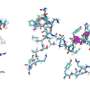
Specifically, it was confirmed that the domain corresponding to half of the glucose transporter proteins located in the N-terminus contained more stable structure formations compared to the proteins from the subsequent C-terminus domain (Figure 2). Using bioinformatics, this characteristic of glucose transporter proteins was found to be conserved in all metazoan sugar transporter proteins (Figure 3). Through these observations, it can be concluded that the cell had to strike a balance in its structure by sacrificing some of its structure-forming ability in order to obtain more functionally superior membrane proteins by developing highly functional chaperones or uniquely-structured lipid molecules.
More information: Hyun-Kyu Choi et al, Evolutionary balance between foldability and functionality of a glucose transporter, Nature Chemical Biology (2022). DOI: 10.1038/s41589-022-01002-w
Journal information: Nature Chemical Biology , Science
Provided by Seoul National University