New software tool allows sensitive deciphering of protein glycosylation
Glycosylation is a common post-translational modification (PTM) of proteins and plays a crucial role in many biological processes. Many disease biomarkers are glycosylated proteins.
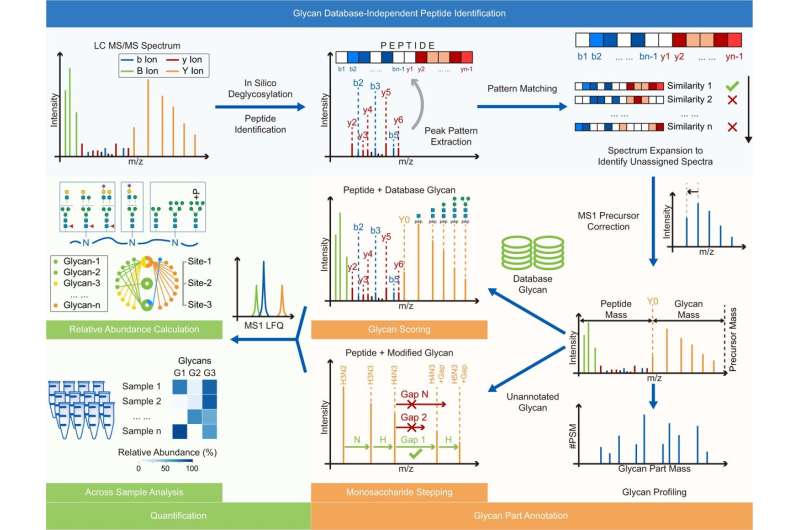
Mass spectrometry-based intact glycopeptide identification can provide information on glycosite and the attached glycan. However, the interpretation of acquired glycopeptide spectra is still challenging.
Recently, a research team led by Prof. Ye Mingliang from the Dalian Institute of Chemical Physics (DICP) of the Chinese Academy of Sciences (CAS) has developed a new glycoproteomics software tool, Glyco-Decipher, for the sensitive interpretation of N-glycopeptide mass spectra at high confidence.
This study was published in Nature Communications on April 7.
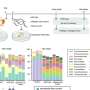
The researchers found that the fragmentation pattern of the peptide backbone of glycopeptides was not affected by the attached glycans or precursor charge states. And the high similarity of the fragmentation pattern of glycopeptides with identical peptide backbones was utilized to interpret glycopeptide spectra with poor peptide fragmentation.
In addition, the scheme of peptide identification in Glyco-Decipher was glycan database-independent, which enabled the discovery of unexpected glycans that were not present in databases. With the developed monosaccharide stepping method, Glyco-Decipher was able to unveil unexpected glycans with modification moieties in complex systems.
Moreover, the quantification module embedded in the software tool extracted elution profiles of glycopeptides, and thus enabled abundance analysis and functional studies of site-specific glycans.
"Glyco-Decipher is overall a very worthy software tool for glycoproteomics," commented one of the reviewers.
More information: Zheng Fang et al, Glyco-Decipher enables glycan database-independent peptide matching and in-depth characterization of site-specific N-glycosylation, Nature Communications (2022). DOI: 10.1038/s41467-022-29530-y
Journal information: Nature Communications
Provided by Chinese Academy of Sciences