Unravelling the coronavirus structure
Since the beginning of the pandemic, scientists worldwide have been working intensely to precisely characterize the novel SARS-CoV2 virus. Only by understanding in detail how the virus is constructed and how it replicates can targets for effective antiviral medications and vaccines be identified. Various Max Planck Institutes are investigating the structure of SARS-CoV2 down to the atomic level, thereby providing valuable insights into the disease. Promising projects from basic research can then be transferred to developing new medications with the help of the Lead Discovery Center.
While vaccinations against SARS-CoV2 have already begun worldwide, there is still no effective medication to treat COVID-19 almost one year into the pandemic. Antiviral medications, which are used for other infections, are not (or only insufficiently) effective. Even the smallest differences in the structure and functioning of viruses have a considerable influence on the effectiveness of medications. Scientists at Max Planck Institutes are therefore closely investigating the surface structures of SARS-CoV2 as well as the processes by which the virus replicates in the infected cell. The aim is to find new targets for possible therapies.
The spike protein is more flexible than previously thought
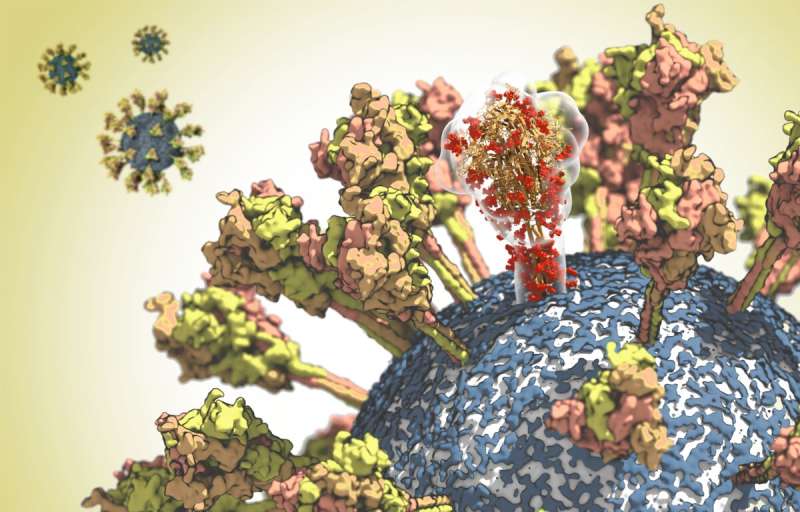
At the Max Planck Institute of Biophysics in Frankfurt, Martin Beck and Gerhard Hummer and their working groups have been studying the structure of the spike protein in detail. The virus needs this surface protein in order to be able to infect cells. The spike protein binds to the ACE2 receptor of human cells. The virus then fuses with the cell membrane and releases its genetic material into the cell interior. However, the exposed location of the spike protein on the viral surface also makes it an important target for the immune system. It is therefore the focus of the development of vaccines and antiviral therapeutics.
With the help of cryo-electron microscopy, Martin Beck has deciphered the structure of the protein at near-atomic resolution. Based on this structural data, Gerhard Hummer then analyzed the properties of the spike protein in its natural environment in computer simulations. The researchers came to some surprising conclusions: The stalk with which the protein is anchored on the surface of the virus proved to be unexpectedly flexible. "The protein probably needs this mobility in order to optimally bind to the receptor on the target cell", explains Hummer. The analyses also showed that antibodies can bind well to the upper part of the spike protein, while other parts of the protein are coated with sugar chains to protect them from being recognized by the immune system. "With this knowledge, we can now identify areas that may be targets for vaccines or therapeutic antibodies", explains Hummer.
Medications on the test bench
The research of Patrick Cramer from the Max Planck Institute for Biophysical Chemistry focuses on the replication of the virus in the cell. Cramer has many years of experience in the study of RNA polymerases, the "copying machines" of the genetic material. "After the outbreak of the pandemic, we developed a unique method to make molecular details visible within a very short time", explains Cramer. For example, he and his team determined the structure of the RNA polymerase of SARS-CoV2 in record time. The precise knowledge of the SARS-CoV2 polymerase now makes it possible to closely study its interaction with antiviral agents.
In an Ebola infection, the medication remdesivir is used to inhibit the viral polymerase. It is the only medication approved in the EU for the treatment of COVID-19. However, it has little effect. Cramer and colleagues showed that remdesivir incorporated into the RNA strand inhibits the continuation of RNA polymerase. However, this inhibition is not permanent. Remdesivir can therefore only slow the replication of the virus but not stop it completely. "We are gaining unique insight into mechanistic details. This, in turn, is giving us a better understanding of the disease", says Cramer. He and his team now want to investigate the interaction of the viral polymerase with other known medications. In cooperation with the Max Planck Institute of Molecular Physiology, the screening of a substance library will help identify completely new active ingredient candidates.
In order to be able to effectively develop promising results from basic research, the Max-Planck-Gesellschaft and Max Planck Innovation have founded the Lead Discovery Center (LDC). Managing Director Bert Klebl would like to close funding gaps between basic research and drug development so that promising projects for combating the coronavirus are not discontinued simply because of a lack of funding.
More information: Hauke S. Hillen et al. Structure of replicating SARS-CoV-2 polymerase, (2020). DOI: 10.1101/2020.04.27.063180 Goran Kokic et al. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir, Nature Communications (2021). DOI: 10.1038/s41467-020-20542-0
Journal information: Nature Communications
Provided by Max Planck Society