Researchers find how enzyme environment alters the rate and direction of a reaction
![Researchers found that subtle changes in the protein scaffold surrounding an enzyme’s catalytic center exquisitely control the rate and direction of chemical reactions. Within [FeFe]-hydrogenases, secondary interactions directly influenced the relative stabilization/destabilization of different oxidation states of the active site metal cluster. Credit: PNNL Researchers find how enzyme environment alters the rate and direction of a reaction](https://scx1.b-cdn.net/csz/news/800a/2020/70-researchersf.jpg)
As researchers look for ways to develop and improve everyday products ranging from fuel to medicines, enzymes provide nature's example of the control needed to make chemical reactions work in desired ways. But a phenomenon known as catalytic bias continues to stump scientists.
The protein scaffold around an enzyme's catalytic core exquisitely controls reactivity, including the direction and rate of chemical processes. Scientists refer to this fine tuning as "catalytic bias"—and how it occurs remains widely debated.
The mystery is one step closer to being solved.
A research team from three U.S. Department of Energy (DOE) national laboratories and four universities found that subtle changes to the environment surrounding some enzymes can not only change the rate of a cellular reaction by a staggering six orders of magnitude but also its direction. That reversal—the root of the catalytic bias dilemma—is like speeding in one direction at 10 miles-per-second, then going in the opposite direction at 1,000,000 miles-per-second. The gap quickly adds up.
"Depending on the desired outcome, the bias could be insurmountable or one of the best things to ever happen," said Simone Raugei, a computational scientist at PNNL and co-author on the study. "A reaction can have many intermediates and can take multiple pathways—some more efficient than others. So being able to tune the catalyst environment can make a remarkable difference in the reaction rate."
The findings, reported in the Journal of the American Chemical Society, hold significant implications for the design of synthetic catalysts used in industrial settings, where predicting and controlling the behavior of a reaction is critical. The results also provide a greater fundamental understanding of the factors that control metabolic processes in all life.
Catalytic bias in metalloenzymes
In nature, the microbe Clostridium pasteurianum (Cp) produces several iron-based enzymes, [FeFe]-hydrogenases, that are among a large group of metalloenzymes. Metalloenzymes are involved in a multitude of reactions that serve many functions, such as moving hydrogen through microbial communities, grabbing nitrogen from the air, producing methane, and converting carbohydrates.
The versatility makes the metalloenzymes, and [FeFe]-hydrogenases specifically, ideal subjects for exploring fundamental questions related to chemical transformations. In fact, hydrogenases have been the source of inspiration for numerous studies of electrocatalysis in energy conversion reactions funded through the DOE's Office of Basic Energy Sciences, said Morris Bullock, director of the Center for Molecular Electrocatalysis, a DOE Energy Frontier Research Center.
"Discoveries reported by different research groups using complementary approaches enrich our understanding of the fundamental scientific basis of catalysis by both biological and molecular systems," said Bullock.
PNNL joint-appointee John Peters is a professor at Washington State University (WSU) and the director of Biological and Electron Transfer and Catalysis, another DOE Energy Frontier Research Center. Peters first started studying [FeFe]-hydrogenases in the late 1990s using enzymes found in the Cp microbe. But those earlier studies focused on general mechanistic aspects, and only recently have researchers started thinking about enzymes in the context of catalytic bias.
Peters and Paul King, manager of the Physical Biochemistry and Photosynthesis Group at the National Renewable Energy Laboratory (NREL), co-designed the new study with Raugei at PNNL and Anne Jones, an associate professor in the School of Molecular Sciences at Arizona State University (ASU).
The study aimed to identify how three [FeFe]-hydrogenases differ in controlling both proton reduction to molecular hydrogen, as well as the reverse reaction—oxidation of molecular hydrogen to protons. Each so-called redox reaction plays a critical role in biological energy transfer and has been extensively studied for energy storage and production applications.
Electrostatics fine tune reaction intermediates
The study of catalytic activity of the three [FeFe] hydrogenases involved a powerful combination of experimental approaches at numerous research facilities and performed by scientists from WSU, NREL and ASU. Raugei, also a joint appointee at WSU, led the theoretical and computational modeling team, whose job was to sort out the experimental observations and develop a theoretical model for the enzymatic mechanism.
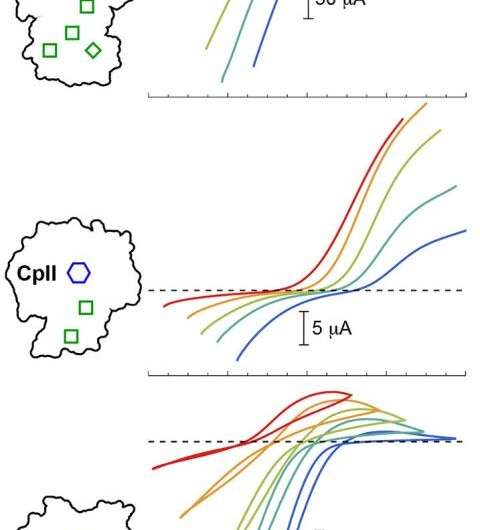
The research team used precise measurements from X-rays at the Linac Coherent Light Source facility, located at the Stanford Synchrotron Radiation Lightsource, to analyze the atomic structure of the enzyme. Next, researchers at ASU subjected the enzyme to an electric current to examine the electrocatalytic activity of each hydrogenase in both resting and active states. All the enzymes shared an identical catalytic cluster of iron and sulfur atoms—generally called the H-Cluster—but differed in the composition of the surrounding protein environment.
The researchers noted significant differences in the reaction bias between the three hydrogenases. Then, during feedback loops between the experiments and computational modeling, subtle changes in the electrostatic field around the catalytic center caught the team's attention.
"We saw indeed that if the field was changed, we were able to drive the reaction more favorably one way or another to try and explain the experimental observation," said Raugei. The subtle differences in the environment, he said, determined the direction of the reaction under steady state conditions and fine-tuned the reaction rate.
According to King, the selective stabilization or destabilization of catalytic intermediates preferentially promoted hydrogen oxidation or proton reduction. Specifically, a more hydrophobic environment surrounding the H-Cluster tuned the enzyme to preferentially favor hydrogen production, whereas hydrogen oxidation was favored by a less hydrophobic environment.
"This work," said King, "reveals the power of biology to control the reactivity of metalloenzymes for meeting the unique metabolic processes in all life and provides valuable new insight for the design of improved electrocatalytic platforms."
Peters said the design features that [FeFe]-hydrogenases use to tune and control reactivity are likely shared among other metal-site biocatalysts. The researchers plan to refine their model to provide a more general blueprint for understanding catalytic bias in the larger class of redox enzymes—and the metabolic energy that drives all life.
More information: Jacob H. Artz et al. Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases, Journal of the American Chemical Society (2019). DOI: 10.1021/jacs.9b08756
Journal information: Journal of the American Chemical Society
Provided by Pacific Northwest National Laboratory