Resurrected protein reveals structure of important enzyme
To disarm toxic substances, many organisms including humans possess enzymes called flavin-containing monooxygenases (FMOs). Despite their importance, the structure of the enzymes has not been resolved, as the protein is too unstable to study in detail. University of Groningen enzyme engineer Marco Fraaije and colleagues from Italy and Argentina reconstructed the ancestral genetic sequences for three FMO genes and subsequently studied the enzymes. The ancestral enzymes proved to be stable enough to study and revealed how FMOs metabolize toxic substances. The results were published on 23 December in Nature Structural & Molecular Biology.
FMOs are present in animals, plants and bacteria. Humans have five FMOs are involved in metabolizing toxic substances and drug activation, while mutations in FMO genes may cause illnesses. "These enzymes are studied in detail by pharmaceutical companies, but we still had no detailed structure available," says Fraaije. Human FMOs are membrane-bound proteins, which proved to be impossible to crystallize for standard structural analysis by X-ray diffraction. "My group published the structure of a bacterial FMO, some fifteen years ago, but this was not a membrane-bound protein."
Phylogenetic tree
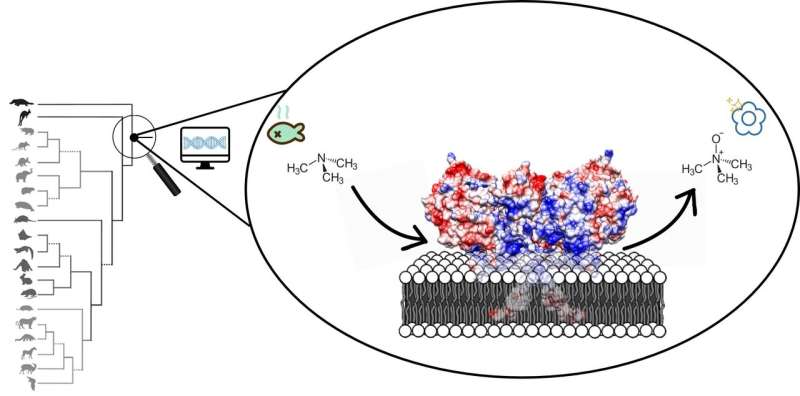
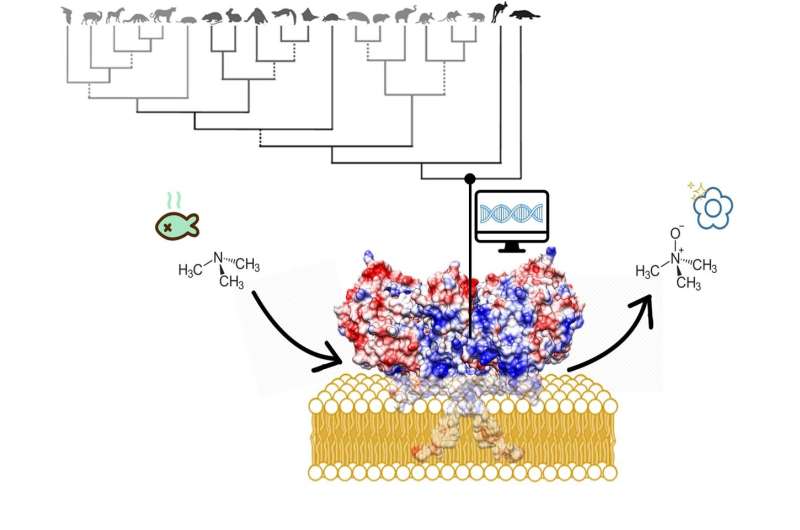
Several developments prompted Fraaije and his co-workers to have another go at the structure: "Over the last couple of years, the gene sequences for a large number of different FMOs were published. This allows you to build a phylogenetic tree, but also to reconstruct ancestral gene sequences for the FMOs." These gene sequences are used to create artificial DNA, which is then translated to proteins in bacterial systems. "Previous work has shown that such ancestral proteins are generally more stable than the modern ones," says Fraaije.
Colleagues from the Universidad Nacional de San Luis in Argentina reconstructed the DNA sequences, the University of Groningen group produced and characterized the proteins, while scientists at the University of Pavia in Italy determined the structures. The international team managed to analyze the ancestral forms of three of the five human FMOs.
Tunnel
"The results are fascinating," says Fraaije. "The membrane-bound part of the enzyme forms a kind of tunnel through which substances can be transported to the active site." Many toxic compounds are fatty substances that will accumulate in the fatty cell membrane. "The FMO enzymes can take them from the membrane and oxidize them." This makes the toxins more hydrophilic, which makes it easier for the cell to excrete them. Whereas the active site is the same in the three FMOs, they have slightly different tunnels, probably suited to different classes of toxic compounds. "We knew that different FMOs metabolize different substances and now we can explain why this is so."
The approach using reconstructed ancestral genes has paid off. "The amino acid sequence of ancestral proteins and contemporary proteins are 90 percent identical, while their functions are completely identical," says Fraaije. Scientists and pharmaceutical companies are now finally able to see how the FMOs work. "This could help in the design of drugs that are activated by these enzymes. And the observation that the ancestral protein is more stable is also of interest. Understanding why this is the case could help us to design more stable enzymes for industrial use."
Fish odor syndrome
Finally, it is now possible to reconstruct the effect of disease-causing mutations in FMO genes. One of those mutations causes fish odor syndrome, where a mutation in FMO3 results in the inability to metabolize the substance trimethylamine. This substance, which has a strong fish odor, consequently builds up in the body and is released in sweat, urine and breath, among other things. Fraaije: "This was a high-risk project, as we didn't know if the ancestral protein would be stable enough. But it has paid off."
More information: Ancestral-sequence reconstruction unveils the structural basis of function in mammalian FMOs, Nature Structural & Molecular Biology (2019). DOI: 10.1038/s41594-019-0347-2 , nature.com/articles/s41594-019-0347-2
Journal information: Nature Structural & Molecular Biology
Provided by University of Groningen