'DNA microscopy' offers entirely new way to image cells
Microscopy just got reinvented—again.
Traditionally, scientists have used light, x-rays, and electrons to peer inside tissues and cells. Today, scientists can trace thread-like fibers of nerves throughout the brain and even watch living mouse embryos conjure the beating cells of a rudimentary heart.
But there's one thing these microscopes can't see: what's happening in cells at the genomic level.
Now, biophysicist Joshua Weinstein and colleagues have invented an unorthodox type of imaging dubbed "DNA microscopy" that can do just that. Instead of relying on light (or any kind of optics at all), the team uses DNA "bar codes" to help pinpoint molecules' relative positions within a sample.
With DNA microscopy, scientists can build a picture of cells and simultaneously amass enormous amounts of genomic information, Weinstein says. "This gives us another layer of biology that we haven't been able to see."
Weinstein, Howard Hughes Medical Institute (HHMI) Investigator Aviv Regev, and molecular biologist Feng Zhang, who was selected as an HHMI investigator in 2018, report the work June 20, 2019, in the journal Cell.
"It's an entirely new category of microscopy," Regev says. "It's not just a new technique, it's a way of doing things that we haven't ever considered doing before."
Something new
Until now, microscopy fit into two main categories. The first is based on optics; light microscopy, for example, dates back to the 1600s and relies on visible light to illuminate samples. Scientists have riffed on this approach, even going beyond the visible spectrum. Electron microscopes, fluorescence microscopes, light-sheet microscopes ¬- they're all based on the principle that samples emit photons or electrons, and the microscope detects the emission.
The second category is based on dissecting samples at locations defined by a microscope. Computer programs then stitch together each dissected piece into a complete picture of the intact sample. Optical imaging can offer intricate portraits of subcellular structure and action. Dissection-based microscopy can give scientists genetic information.
Weinstein and his colleagues at the Massachusetts Institute of Technology wanted to create a way to do it all in one shot—to take a snapshot of a cell's position and spell out the specific genetic sequences driving it.
That combo is important for scientists studying genetically diverse sets of cells. The immune system is a perfect example, Weinstein says. Immune cell genes can vary down to a single letter of DNA. Each variation can trigger a dramatic shift in the type of antibodies a cell produces. Where that cell is located within a tissue can alter antibody production, too.
If you focus on just one or the other, "you're only getting part of the picture," he says.
How it works
Capturing such a complete picture of a cell doesn't require an expensive microscope or a lot of fancy equipment, Regev says. All you need to get started is a specimen and a pipette.
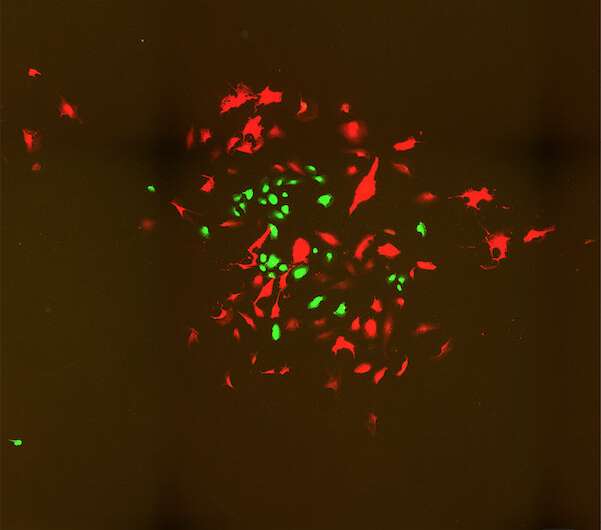
First, scientists take cells grown in the lab and fix them into position in a reaction chamber. Then, they add an assortment of DNA bar codes. These stick to RNA molecules, giving each a unique tag. Next, the team uses a chemical reaction to make more and more copies of each tagged molecule—a growing pile that expands out from each molecule's original location.
"Picture every single molecule as a radio tower broadcasting its own signal outward," Weinstein says.
Eventually, the tagged molecules collide with other tagged molecules, forcing them to link together in pairs. Molecules located close to one another will be more likely to collide, generating more DNA pairs. Molecules further apart will generate fewer pairs.
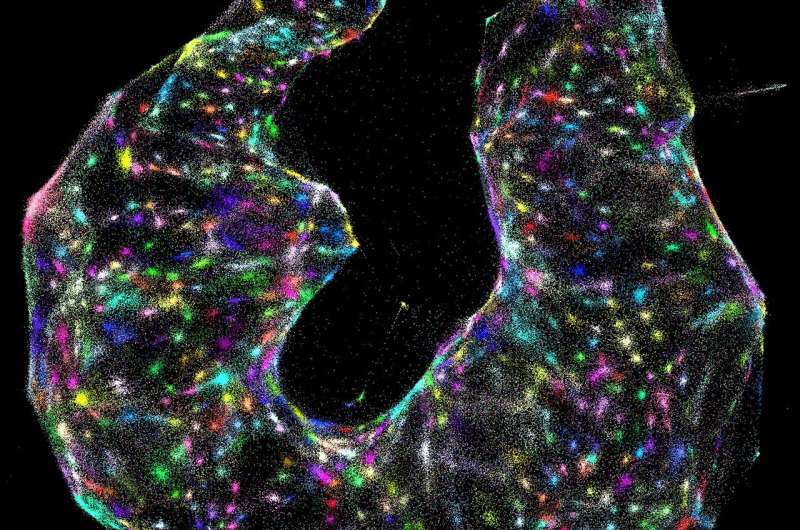
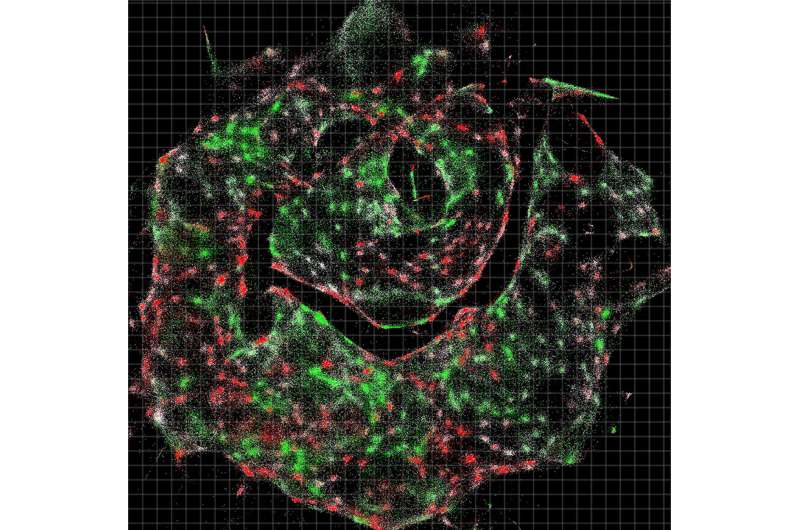
A DNA-sequencing machine spells out the letters of every molecule within the sample, which takes up to 30 hours. An algorithm the team created then decodes the data—which, in the paper, represents roughly 50 million DNA letters of genetic sequences from each original specimen—and converts the raw data to images.
"You're basically able to reconstruct exactly what you see under a light microscope," Weinstein says.
The two methods are complementary, he adds. Light microscopy can see molecules well even when they're sparse within a sample, and DNA microscopy excels when molecules are dense—even piled up on top of one another.
He thinks DNA microscopy could one day let scientists speed the development of immunotherapy treatments that help patients' immune systems fight cancer. The method could potentially identify the immune cells best suited to target a particular cancer cell, he says.
Every cell has a unique makeup of DNA letters, or genotype, Zhang says. "By capturing information directly from the molecules being studied, DNA microscopy opens up a new way of connecting genotype to phenotype."
The possibilities with this category of microscopy are wide open, Regev adds. "We hope that it sparks the imagination—that people will be inspired with great ideas that we've never thought about."
More information: Joshua A. Weinstein et al. "DNA microscopy: Optics-free spatio-genetic imaging by a stand-alone chemical reaction." Cell. Published online June 20, 2019. DOI: 10.1016/j.cell.2019.05.019
Journal information: Cell
Provided by Howard Hughes Medical Institute



















